

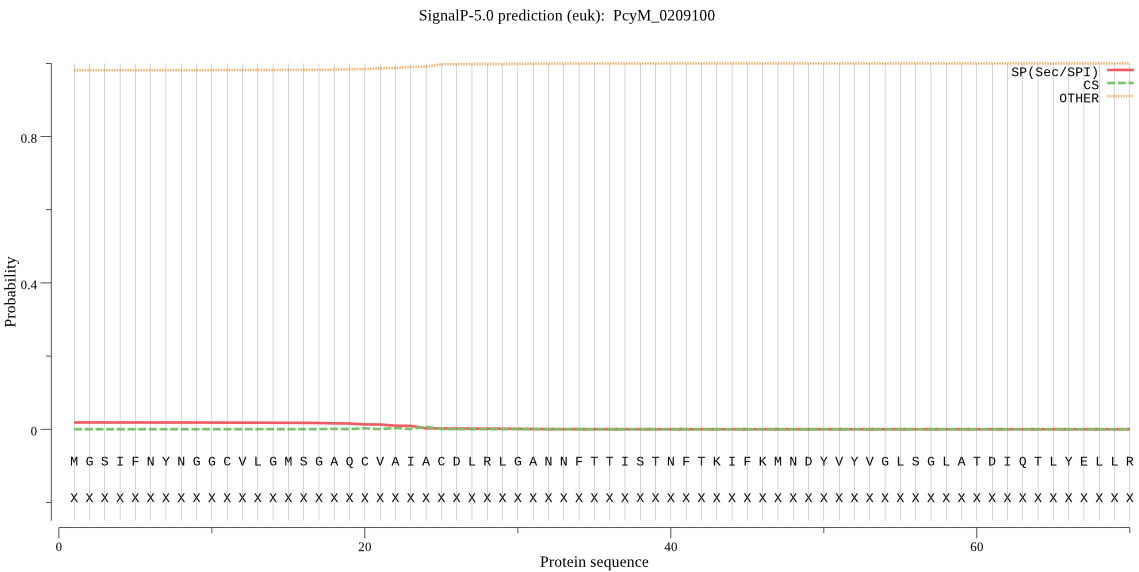
| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0209100 | OTHER | 0.926308 | 0.063294 | 0.010398 |
MGSIFNYNGGCVLGMSGAQCVAIACDLRLGANNFTTISTNFTKIFKMNDYVYVGLSGLAT DIQTLYELLRYRVNLYEIRQESPMDIDCFANMLSSILYANRFSPYFVNPIVVGFRVKHTI DDEGKKISTFEPYLTAYDLIGAKCETNDFVVNGVTGEQLYGMCESLYIKDQDKDGLFETI SQCLLSALDRDCLSGWGAEVYVLTPDTIMKKKLKARMD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0209100 | 82 S | IRQESPMDI | 0.998 | unsp |