

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0216600 | OTHER | 0.999294 | 0.000145 | 0.000561 |
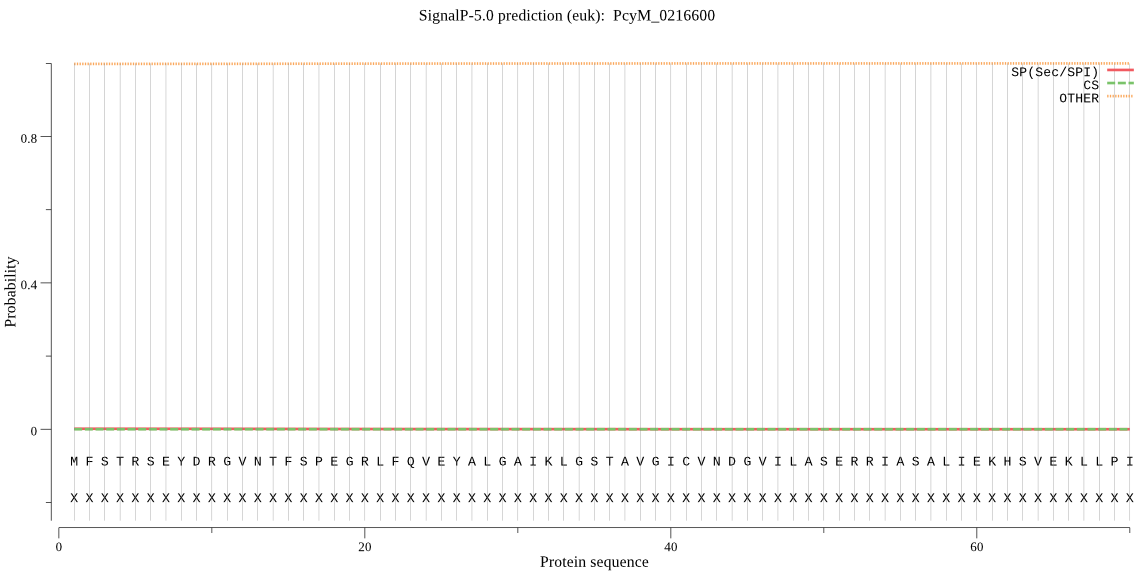
MFSTRSEYDRGVNTFSPEGRLFQVEYALGAIKLGSTAVGICVNDGVILASERRIASALIE KHSVEKLLPIDDHIGCAMSGLMADARTLIDYARVECNHYKFIYNENINIKSCVELISELA LDFSNLSDNKRKKIMSRPFGVALLIGGVDKNGPCLWYTEPSGTNTRFLAASIGSAQEGAE LLLQENYNKDMTFEEAEILALTVLRQVMEDKLSSSNVEIAAVKKSDQTFYKYNTEDISRI IEALPSPIYPTIDMTA