

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
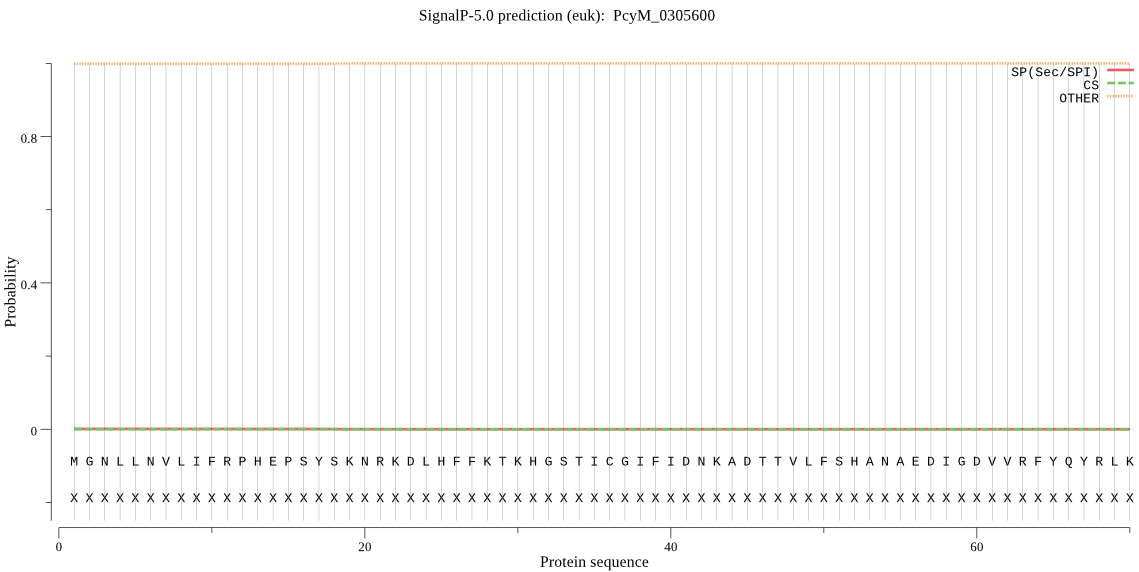
| PcyM_0305600 | OTHER | 0.998730 | 0.000711 | 0.000559 |
MGNLLNVLIFRPHEPSYSKNRKDLHFFKTKHGSTICGIFIDNKADTTVLFSHANAEDIGD VVRFYQYRLKRLGLNLFAYDYSGYGHSSGYPTETHLYNDVEAAYDYLVTELRIPRNSIIA YGRSLGTAASVHIATKNNLLGVILQAPLASIHRVKLKVKYTLPYDSFCNIDKVHMIKCPI LFIHGTKDRLLSYHGTEEMIRRTTVNTYYMFIQGGGHNDLDCSFGSLVNAALVAFLFVLK HNIRENVNSVYNISDVNIQKLRNKFILNNAKMVKEKLKEKHRKGEEDTEMLSVSTENLNL GPIDRNKASGSSEVLNSFMSNMENWYIEQSRPWDKRKKAGYDSSRSVSTVLGSAESSMKN IPDNTFYESDTNVSIEDLWRYTCAKYNESERKLHGGRDSGGVSSSSFTKQRGAKITNRAF ICELSEGSDRRGYRERSRERYPDGRRIDSRERYPDGRRIDSRERYPDGRRIDSRERYPDG RRIDSRERHPDGRRIGSREEGMERIRIKGCSSYVYNPARRENMPPSHMDQSPSHSSRSNI HMGSEKDLAQRGKGSAMDDSSSEKINSRMSRESRNVYIVDKVPRVSQDMKNAVVPPVDER GSGSLHGAHASYAHSDEECGNEREKAIGAHESGSEYSGESRRGEGEGKGKGKGKGKGKGK GKGGMSSGMAEVEIGSHSPHERVIQGMHSDNDRALEAMGGGDISASKVSLSSVSSNESKA SSMSSGVSVESRVSSSCGASKEVRVEGMIESRRNMESRRNMESNAAVQGRSDDGGGSNTS EQVCAKSYYMKRGNNIRSEMRNKITDMNNLKFNSYNDFIYGNQENKMDEYPNQAMEPIGR KMSSASSSTMNNLSSLFGYRSISNEKLG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0305600 | 374 S | DTNVSIEDL | 0.993 | unsp | PcyM_0305600 | 374 S | DTNVSIEDL | 0.993 | unsp | PcyM_0305600 | 374 S | DTNVSIEDL | 0.993 | unsp | PcyM_0305600 | 437 S | YRERSRERY | 0.997 | unsp | PcyM_0305600 | 449 S | RRIDSRERY | 0.996 | unsp | PcyM_0305600 | 461 S | RRIDSRERY | 0.996 | unsp | PcyM_0305600 | 473 S | RRIDSRERY | 0.996 | unsp | PcyM_0305600 | 485 S | RRIDSRERH | 0.997 | unsp | PcyM_0305600 | 497 S | RRIGSREEG | 0.997 | unsp | PcyM_0305600 | 526 S | NMPPSHMDQ | 0.993 | unsp | PcyM_0305600 | 544 S | IHMGSEKDL | 0.99 | unsp | PcyM_0305600 | 570 S | NSRMSRESR | 0.997 | unsp | PcyM_0305600 | 678 S | IGSHSPHER | 0.995 | unsp | PcyM_0305600 | 714 S | LSSVSSNES | 0.99 | unsp | PcyM_0305600 | 722 S | SKASSMSSG | 0.992 | unsp | PcyM_0305600 | 728 S | SSGVSVESR | 0.991 | unsp | PcyM_0305600 | 734 S | ESRVSSSCG | 0.992 | unsp | PcyM_0305600 | 192 S | DRLLSYHGT | 0.995 | unsp | PcyM_0305600 | 357 S | SAESSMKNI | 0.99 | unsp |