

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0306800 | SP | 0.000950 | 0.999046 | 0.000004 | CS pos: 17-18. AKC-DI. Pr: 0.8671 |
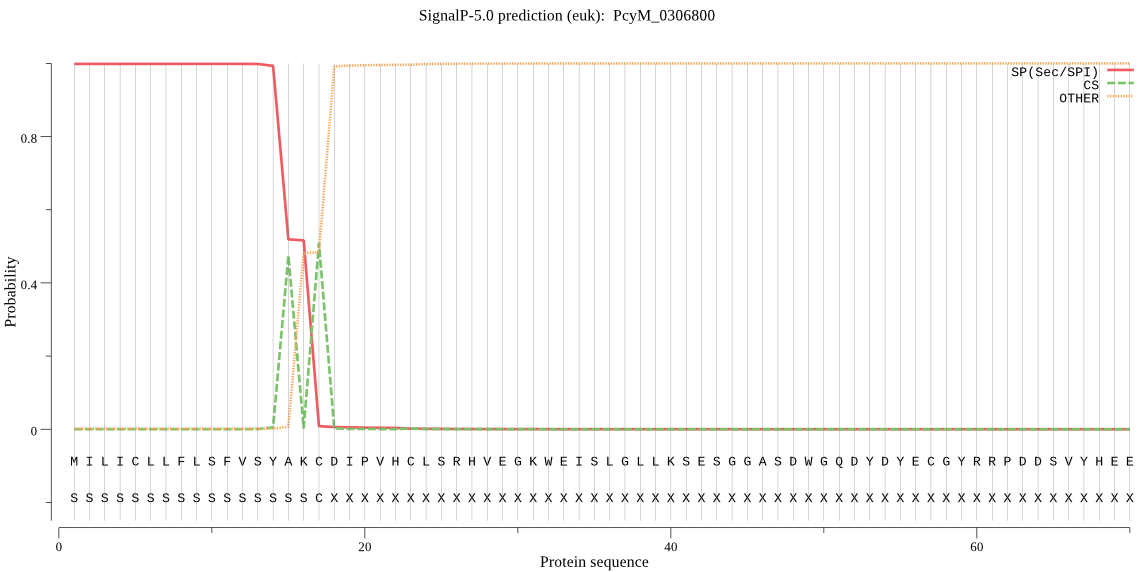
MILICLLFLSFVSYAKCDIPVHCLSRHVEGKWEISLGLLKSESGGASDWGQDYDYECGYR RPDDSVYHEELDPDHLKRRFEEKKKQIIQFNTDRTINIIENGDINPEYSGFWRIVYDEGL YMEIYKQNEEKEIYFSFFKFQRRNDTSYSYCNRLIMGVVNVYHLNGNEGGSSSGLPRIED FPDQNAGGEKGPKLGDVFLVKKEHKRTNGLNPNRINLSKESFHYVDSKEGARKSSSHSID YYNDNYIQFDFFTMKRFCWYGKKVGTPDEPTNKIPVEIISPLVLDPDTHNKNYANVKLEW GGSNGGGTQRVDHEEVGTDHIVTQSRDPSKHTNKSHKGGKQNSIFNTYREKEVTLAQFDW TNEADVRKRLNGNWVKVIDEAIDQKNCGSCYANSAALIINSRVRIKYNYIKNIDLLSFSN KQLVLCDFFNQGCNGGYIYLSLKYAYENFLFTNKCFVRYSNLYLNRDKKNSALCDRFDTF KIFLQKGRGGNFSNAQVGPPHRQTKRVRQHEPFRGSEEVVRSEEVDRGEEVVRSEEVGRG EEVVRSEEVGRGEDVGKSPTQGEQHKGGIFHKGENKHSGRHMHHMHHTRDVRDVRGGREL PRDDQLNEGYLPVDASARDASELDVLKLHECDAKVKVTKFEYLDIEDEEDLKKYLYHNGP VAAAIEPSKNFSAYREGILTGKFIKMSDGGESNAYVWNKVDHAVVIVGWGEDTVENLKKK KKKIHHYSGHPGAYASGKGGAAAGKVVKYWKILNSWGTHWGYDGYFYILRDENYFSIKSY LLACDVSLFIRRGDSRVTPG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0306800 | 504 T | PHRQTKRVR | 0.994 | unsp | PcyM_0306800 | 504 T | PHRQTKRVR | 0.994 | unsp | PcyM_0306800 | 504 T | PHRQTKRVR | 0.994 | unsp | PcyM_0306800 | 558 S | DVGKSPTQG | 0.992 | unsp | PcyM_0306800 | 65 S | RPDDSVYHE | 0.99 | unsp | PcyM_0306800 | 335 S | HTNKSHKGG | 0.996 | unsp |