

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
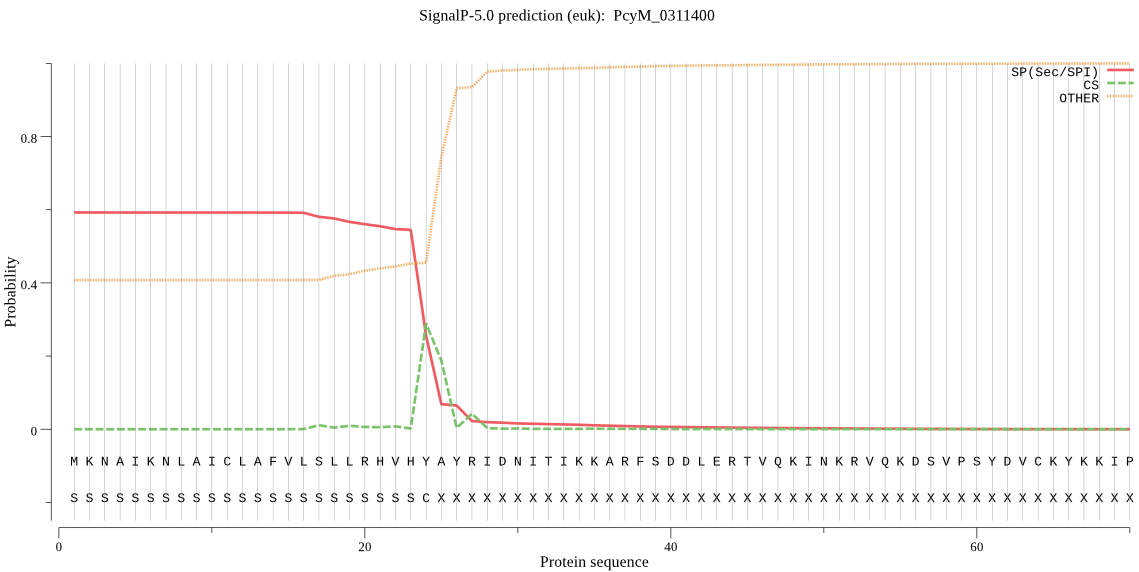
| PcyM_0311400 | SP | 0.183890 | 0.811259 | 0.004851 | CS pos: 24-25. VHY-AY. Pr: 0.4322 |
MKNAIKNLAICLAFVLSLLRHVHYAYRIDNITIKKARFSDDLERTVQKINKRVQKDSVPS YDVCKYKKIPKFLTERNKLKDTQKVKYIVGIENTCDDTCICVLDSNLNIVKNVIISHFKV VHKYEGVYPFFVSSINQLFLKPYVEKTLEGIDESRISCFAFSACPGIAKSMEAAKNYIGE KKKQNGSIQVSPINHVFAHVLSPLFFHVYNDKNTYTNSGQYKEGKSEQSKNELGIDMHIS EEVRKDKTQMEKVKGILQILNSNDVTKRKMALLSEEEFLNCGNYVLRGEMGNETPQSGDS EELQSEESEALQNVESASPQSVESASLQRAESEAPQNVAYQRKNTSSAQKTQYSGDAPQG SHLTDGYICALVSGGSTQIYKVQRNKKNDINLCKLSQTVDISVGDIIDKIARLLGLPVGL GGGPFLERKSAEFVTRMKEERLSGEACADPFQPFPVPFSANSRIDFSFSGLYNHLRKIIE ESKKGESFEREKDRYAYYSQKNIFNHLLKQMNKIMYFSELHFNIKKLFIVGGVGCNHFLI TKLKSMAMKRSQLQVQLNEFKKLKKRLSKRVKKISEKKFLHFKVSKEVLKGEIEFTASLG WDIYLKLLLKRKTERDILSALKYFNFEDFSKLKEGGHFLLEGHSASSKSEAPWSVHKTPP NLSRDNAAMICFSAFLNLHNKSDLYEDVSQVKIKPTVKTQVENNFLLFSDMIIFDVFLQQ FGAQGG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0311400 | 443 S | EERLSGEAC | 0.995 | unsp | PcyM_0311400 | 443 S | EERLSGEAC | 0.995 | unsp | PcyM_0311400 | 443 S | EERLSGEAC | 0.995 | unsp | PcyM_0311400 | 487 S | KKGESFERE | 0.996 | unsp | PcyM_0311400 | 568 S | KKRLSKRVK | 0.995 | unsp | PcyM_0311400 | 575 S | VKKISEKKF | 0.993 | unsp | PcyM_0311400 | 647 S | HSASSKSEA | 0.997 | unsp | PcyM_0311400 | 300 S | QSGDSEELQ | 0.991 | unsp | PcyM_0311400 | 321 S | ASPQSVESA | 0.992 | unsp |