

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
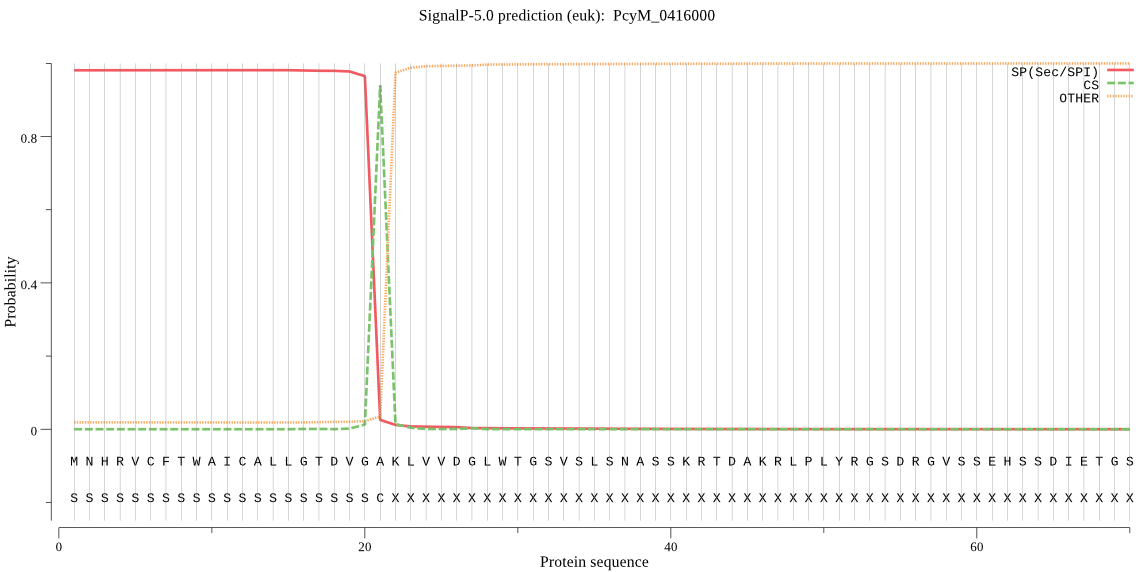
| PcyM_0416000 | SP | 0.019603 | 0.980241 | 0.000156 | CS pos: 21-22. VGA-KL. Pr: 0.9593 |
MNHRVCFTWAICALLGTDVGAKLVVDGLWTGSVSLSNASSKRTDAKRLPLYRGSDRGVSS EHSSDIETGSAVPHPAPNVAPRNADDAAGGGQPPQEVDHPLNRVTSVMAAPVSKRSEVEV RSALLKNHDGVKITGTCNAKFQLFLVPHISISVEAESNTIQLGRKLEVVTITKKQHKVVG SKSSPLLQFEEDEKFLLNQCAEGKAFKFVVIVKGEELILKWKVYEREPSATDNNKVDVRK YVLRNLGSPITSIQVHSGKEDNDVFLLESKAYLLRQDIPKTCERIATNCFLSGNVDIEGC FKCTLLWENTKLGSPCFSYLPPDVKHNYEQIKMKAQDEGDKEEAQLDEFIRRVLQMVQKM EKNNTPRKGKNKGDYPNDNLKELLLSYCQMMKKVDTSGTLEEHELGNEVDVLTNLEGLLR KHSNEEIAVLREKLKNPAICMKDADMWIVQKRGLALPTFEYKHLQRRRVTTPVDSKEDNK ETDRRNVIKDMYIPGYRAVIDLSQKSEMNHSNPSGEMFCNEDYCDRWKDKNSCFSSIETE EQGNCNLSWLFASKTHLETIRCMKGYDHLGSSALYVANCSKRGNKSKCTSGSNPYEFLTI VEENGFLPPALLIPYSYADVGNGCPRKENHWQNLWANVKLLEPSDEPNSVSTKGYTSYES DDFRGNIDVFIDLVKREVRGKGSVMAYVKALGVLGYNMNGKEVHSLCGDKRPDHAVNIIG YGNYINTQGLKKSYWLVRNSWGKYWGDEGNFKVDMHGPADCQHNFIHTAAVFNLHVPMSE GPSKREAHLYNYYLKSSPDFLGNIYYKNIRRMSGVAEKGRTGRHQSLAVQGQEGQSDLLE GSTPSGTANQVDEPKGAREDAPKRGDQGKCMSEDPPIEVVNEELISESTLTLIPQAARRE GQVEANPAPPLTVAASLPLRGDVLLVRDGTDEGSTEAATVEVLHILKHIKHGKVKLGIVT YGDESDISGDHSCSRSLAQDSEQLTECIQFCHNEWPNCRDEASPGYCLAQRRKTGDCFFC YV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0416000 | 64 S | SEHSSDIET | 0.992 | unsp | PcyM_0416000 | 64 S | SEHSSDIET | 0.992 | unsp | PcyM_0416000 | 64 S | SEHSSDIET | 0.992 | unsp | PcyM_0416000 | 229 S | EREPSATDN | 0.992 | unsp | PcyM_0416000 | 257 S | IQVHSGKED | 0.994 | unsp | PcyM_0416000 | 423 S | LRKHSNEEI | 0.996 | unsp | PcyM_0416000 | 471 T | RRVTTPVDS | 0.995 | unsp | PcyM_0416000 | 475 S | TPVDSKEDN | 0.998 | unsp | PcyM_0416000 | 535 S | NSCFSSIET | 0.995 | unsp | PcyM_0416000 | 657 S | KGYTSYESD | 0.997 | unsp | PcyM_0416000 | 783 S | SEGPSKREA | 0.996 | unsp | PcyM_0416000 | 813 S | IRRMSGVAE | 0.995 | unsp | PcyM_0416000 | 965 S | YGDESDISG | 0.991 | unsp | PcyM_0416000 | 39 S | LSNASSKRT | 0.997 | unsp | PcyM_0416000 | 59 S | DRGVSSEHS | 0.996 | unsp |