

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
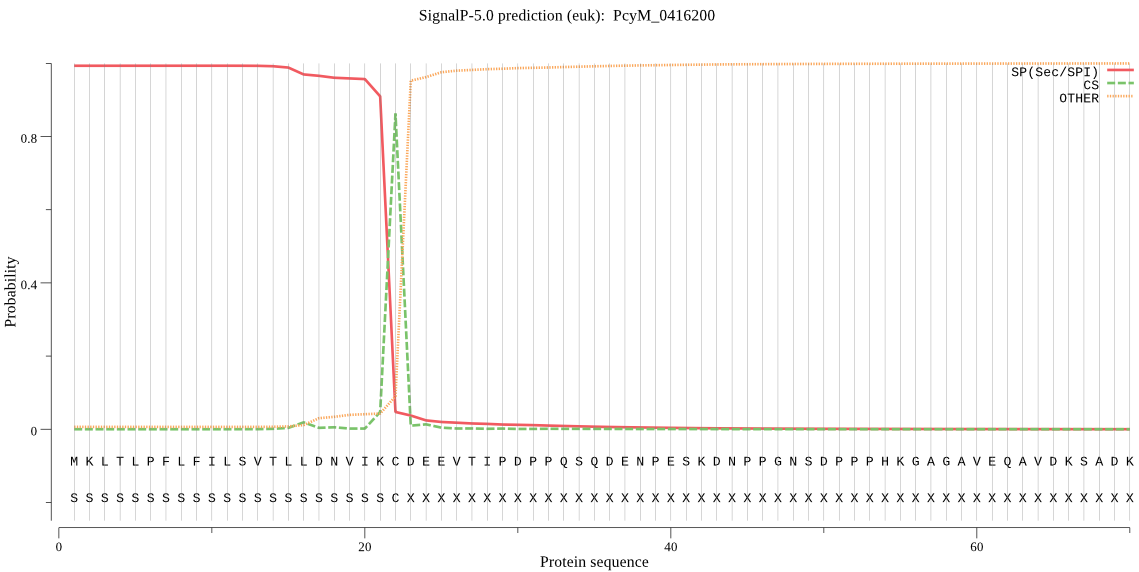
| PcyM_0416200 | SP | 0.004031 | 0.995969 | 0.000000 | CS pos: 22-23. IKC-DE. Pr: 0.9038 |
MKLTLPFLFILSVTLLDNVIKCDEEVTIPDPPQSQDENPESKDNPPGNSDPPPHKGAGAV EQAVDKSADKPVDKPADKPADKPADKPADKPADKPADKPADKPADKPADKPADEPADKPA DKPADEPADQPTDEPTDEPADKPADKPADEPADKPADKPADQPTDQPTDKPADKPADQPT DQPTDQALTQPADQPADQPTEQPTEQPTEQPADQPTDQPTEQPAEQPTEQPADQPTDQPT EQPLTQPNVEASDRATAAALKNPNEIEAQCAQLKDQDGVKITGLCRAKFQVFLVPHVTIN VETETNTIYIGKKLDDVVITKKQNKVVSSKSSPLLQFEENADSLLNQCAEGKTFKFVVIV KGEELILKWKVYEKEPSATDNNKVDVRTFLLKNTDRPITAIQVHTAKGNEESFLLESKSY FLIDDMPAKCGLIATNCFLSGSLDIEGCYKCTILSENTELDSPCFSYLPPDVKHNYEQIK SKAHQVDQKEVQFAVSIGKILQGVYKKGETNLNQLLTFDEADAALKAELLNYCASMKEVD SSGVLENYELGSEEEVFANLTNILKNHAGETKSTLQTKLKNPAICLKNADEWVKSKKGLL LPSLSYTHVEATLPATAPENEEKKEDTPKGSEKIQTNGYDGVIDFVSREETNMQSTSFID NMYCNVEYCNRWKDPSSCMAKIEAGDQGDCATSWLFASKVHLETIKCMKGHDHIASSALY VANCSSKEAKDKCQAPSNPLDFLNTLEETKFLPAESDLPYSYKAVNNVCPEPKNHWQNLW ADVKLLDKQYQPNSVSTKGYTAYQSDHFKGNMDAFIKLVKSEVMNKGSVIAYVKAAGALS YDLNGKKVLSLCGGETPDLAVNIVGYGNYINGEGVKKSYWLLRNSWGKHWGEDGNFKVDM DSPPGCQHNFIHTAAVFNVDLPLLENSEKKRPMLYNYYMKSSPDFYNHILYRGVQTEERE IGISGEDKTSSFVVSGQNSGGDTLDGAEQSSVVVEGKEKPAEGGDESTQEVESPPKAGTD ESKEADEQDEEVEEEEPEEEGDGEQEEERDDESEEEEEDEEQEEGVAEQEVGGADAEVGV AEQEVGGAEAEEGGAEAEEGEAEVGEAEVGGAEAEVGRAEPEEGDSEAAKKGVEEPQTVA SPSASVNSTPPSTEPAPKHNTSLIKVKQIMEVVHIIKHIKNGKVRFGITTYEDDMGIANK HDCSRSYSQDPEKLAECIQFCHDEWNNCKGEPSPGRENDSIYCVDTAFTLAYQWERVPNV YLTLQARRGSFSLLFAQLAKGVVLLYLSFFWIFLIFFDFFDFFNFLIFLIFLIF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0416200 | 627 T | KKEDTPKGS | 0.992 | unsp | PcyM_0416200 | 627 T | KKEDTPKGS | 0.992 | unsp | PcyM_0416200 | 627 T | KKEDTPKGS | 0.992 | unsp | PcyM_0416200 | 964 S | EIGISGEDK | 0.996 | unsp | PcyM_0416200 | 979 S | SGQNSGGDT | 0.994 | unsp | PcyM_0416200 | 1053 S | RDDESEEEE | 0.998 | unsp | PcyM_0416200 | 34 S | DPPQSQDEN | 0.99 | unsp | PcyM_0416200 | 328 S | NKVVSSKSS | 0.991 | unsp |