

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
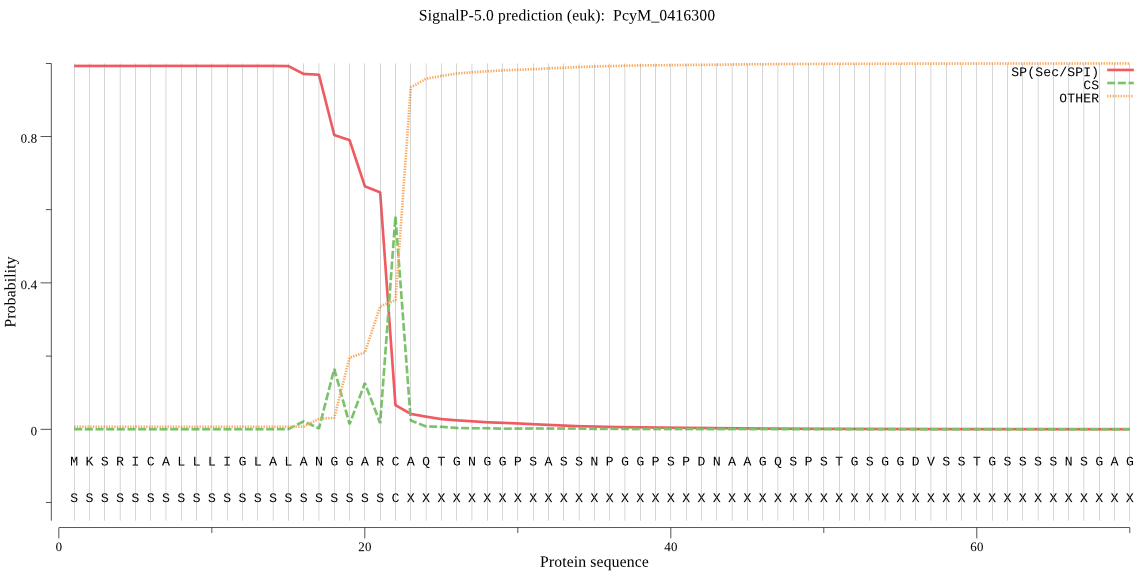
| PcyM_0416300 | SP | 0.001960 | 0.998033 | 0.000006 | CS pos: 22-23. ARC-AQ. Pr: 0.5124 |
MKSRICALLLIGLALANGGARCAQTGNGGPSASSNPGGPSPDNAAGQSPSTGSGGDVSST GSSSSNSGAGGGSDPAAEASRADGSPSPQENNLNIPSPVPQPSNIPSAASAPSAPNAASD ASASSAASADPPEAAPAPAITTNPIEVKSSLLKDQKGLKITGPCKSFFQVYLVPYLYMNV NAERSEIEMEPLFMKVDNRIKFEKEKHLLQNICADNKTFKLVLYIYEGELTIKWKVYPPK GESTNDKMVDIRKYKMKDIGQPITSMQVMVMTEKHKTIYVESKNFSVMNDIPEKCDAIAN ECFMSGVLDIQKCYHCTLLLQKKESAQECFNFVSPEIRSRFDDIQAKGEDEENPNEVELE GTIDKIFIKIYKKGENHYKEVDQLAIMDSSFKSELLKYCSLMKETDSSGALDNHELGNAE EVFAHITTMLQSNSDLNVYSLKNKLKNPAICLKKVGHWIGSKTGLVLPILEHSSVHDSAN TYVETEGSTSNSATTQSADVHLLHVSDKLFCNADYCDWTKDTSSCIAKIEAQDQGDCATS WLFASKVHLETIKCVKGYEHVPSSALYVANCSSKEAKDKCQAASNPLEFLNILEETKFLP AESDLPYSYKAVNNVCPEPKSHWQNLWENVKLLDKQYQPNSVSTKGYTAYQSDHFKGNMD AFIKLVKSEVMNKGSVIAYVKAQGVMSYDLNGKKVLSLCGGETPDLAVNIVGYGNYINGE GVKKSYWLLQNSWGKHWGDKGYFKVDMHGPDHCQKNFIHTAAVFNVDIPVVIPAPNSDPE INNYYLKKSPDFFSNFYFNKLEAESEGNSGGAKSASNNLTVQGQEGSTEESTSVQQSQNG GGESGQSAARGADGQGQSEAVAQVQEGGAVVSSQPQADTIPALPTNGVPSAEASGQQNGE AGQSVSNTLTETQNSSVTTETTEQTTQQNTQQTAQVPSAQSARAPLSTLTGSTGVIVTEV KEALHFLKNVKNGKVKSNFVSYDNADALGDEKVCSRAFSTDVDKQAECIEFCEKNWGACK GKVSPGYCLTKKRGSNDCFFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0416300 | 1035 S | KKRGSNDCF | 0.993 | unsp | PcyM_0416300 | 1035 S | KKRGSNDCF | 0.993 | unsp | PcyM_0416300 | 1035 S | KKRGSNDCF | 0.993 | unsp | PcyM_0416300 | 85 S | RADGSPSPQ | 0.99 | unsp | PcyM_0416300 | 474 S | LEHSSVHDS | 0.992 | unsp |