

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
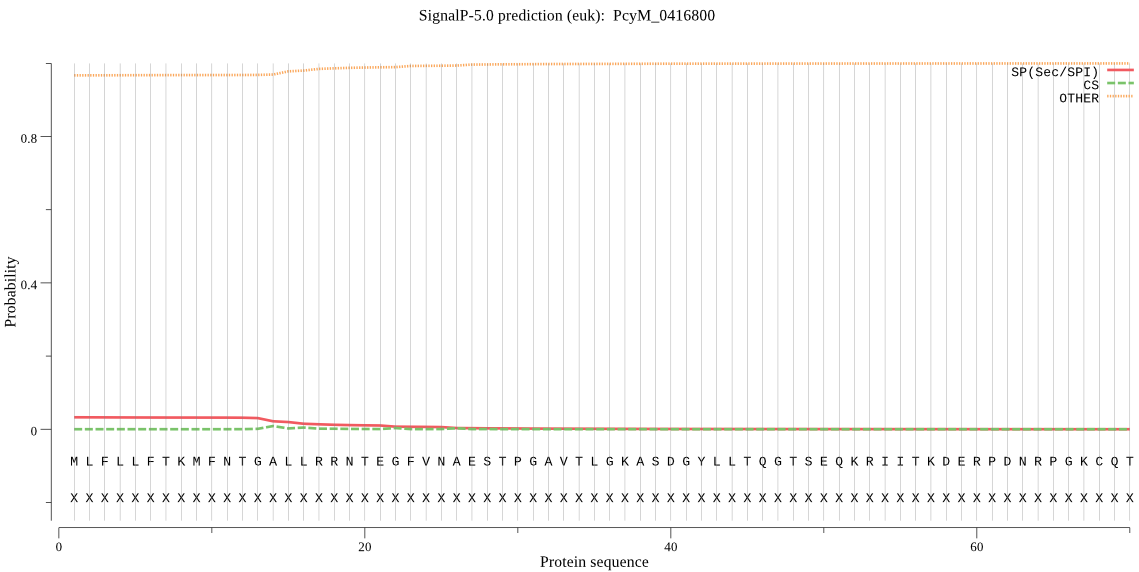
| PcyM_0416800 | OTHER | 0.677920 | 0.113883 | 0.208197 |
MLFLLFTKMFNTGALLRRNTEGFVNAESTPGAVTLGKASDGYLLTQGTSEQKRIITKDER PDNRPGKCQTRHDPTPTTHRTSSNPRRQSVGVSQSNYAKVKAQVLKKTTEVKINRTCGSE MGFLFIPHSYIYVTTKDSKAGAFKMLPHIEDNVLPLSWKAYLPEDPFTATKRKETETKWY RRTLAESNDGPCDEDHDLFSSGIDSEEDEAIFPMEKDDQSEGCSEGYSEGCSEDASEEHQ LRNSIDRVLKGVYKAAEDGRKVLITKEELDSSMKEELIKYCQLLKKVDTSGTLEEHQMGS EMDVFDNLVRLLLKHRDETVFTLRRKLRNPAVCMKNVGEWVLKRRGLALPDS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0416800 | 205 S | SGIDSEEDE | 0.997 | unsp | PcyM_0416800 | 272 S | ELDSSMKEE | 0.997 | unsp |