

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
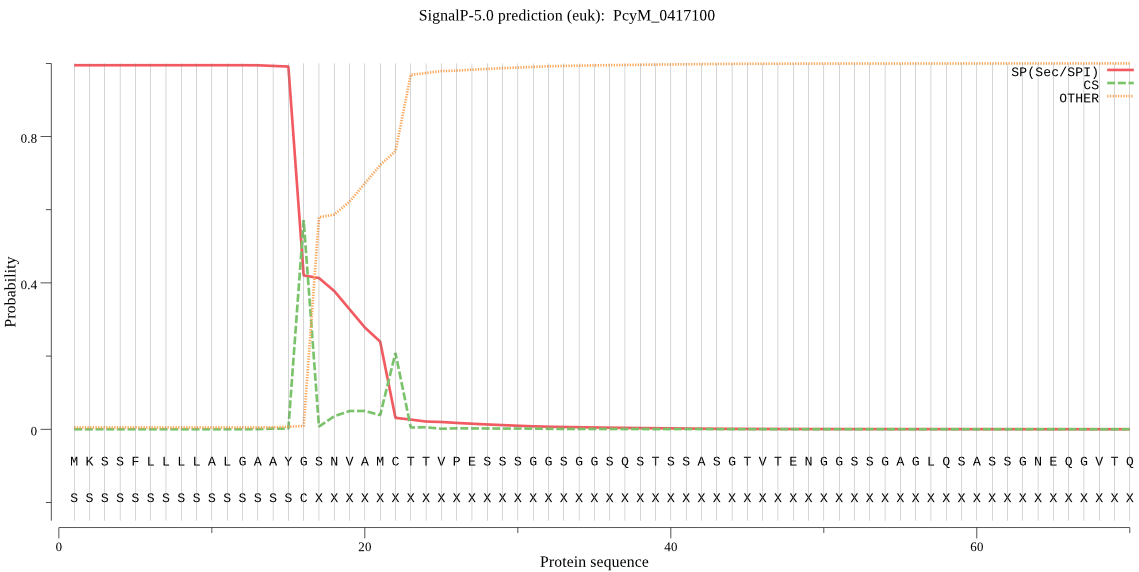
| PcyM_0417100 | SP | 0.000144 | 0.999856 | 0.000000 | CS pos: 16-17. AYG-SN. Pr: 0.6587 |
MKSSFLLLLALGAAYGSNVAMCTTVPESSSGGSGGSQSTSSASGTVTENGGSSGAGLQSA SSGNEQGVTQPTNPSPPVSQAPNSAESNPSQPVQTPDQHADVPPSQSSTTPTAQGIASAS SSGNTNGQGGATQLQTSTKKAELQSALLKNFTGVKVTGPCDTEVGLFLIPHMYISVKAAK DIIELSTKFPNSDNTIIQFTKGGETLINKCEGNPEKTFMFIVYLEDNILTLKWIVYPQSE DKNKADVRKYRLPNLERPITSIQVHSVMVQEDTVIYTSKDYSIKNDIPGESRELGPTDGC SPEQNCQQTMSACFLSGNTDIESCYTCNLLIHNNDTNDKCFDYVSADFKKEFLDIKVKGQ DDEESSEYKLAQVINEVLNGIYKTDPEGNKELITSEELDENVKKHIGNYCQVLKEMDISG TMEVHQMGNEMDVFKNLVQLLQKHGEEKNSTLEWKLQNPALCLKNVNDWVVNKKGLVLPL LQNGGSDIYFGESNLVEGGQKSGLSTYQVGDDGIIDLSIEQKNSHTSSTPFTNHMFCNAD YCDWTKDSSSCMAKIEAGDQGDCATSWLFASKVHLETIKCMKGYDHIASSALYVANCSSK EAKDKCQAPSNPLEFLNILEGTKFLPAESDLPYSYKEVNNVCPEPKSHWKNLWENVKLLD KQYQPNSVSTKGYIAYQSDHFKGNMDAFIKLVKSEVMNKGSVIAYVKADELMGYDLNGKK VLSLCGGETPDLAVNIIGYGNYIHGEGVKKSYWLLQNSWGKHWGDKGKFKVDMDGLPGCQ HNFIHTAAVFNLDVPPFQSPAKDTELYSYQLKSSPDFYKNLYYNAVGEAKGSAPSQAVHG QDAPEETAVGGGENSVSTVQGQQPQPQVPSGVSTDTKAPEGADNTVSTSEISVGPSQQAQ APQSPGEGQVVSTQQQPGGSTPAPEPSAPQPAQQVDASTPVATVTPGGANPNGGAENAKM SHIIHVLKHIKQTKMVTRMVTYEGEYELGDHSCSRTQASTVEKLDECIRFCNENLYLCER TVSAGYCLTKLRNANDCIFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0417100 | 137 S | QLQTSTKKA | 0.997 | unsp | PcyM_0417100 | 137 S | QLQTSTKKA | 0.997 | unsp | PcyM_0417100 | 137 S | QLQTSTKKA | 0.997 | unsp | PcyM_0417100 | 282 S | SKDYSIKND | 0.996 | unsp | PcyM_0417100 | 904 S | QAPQSPGEG | 0.995 | unsp | PcyM_0417100 | 28 S | TVPESSSGG | 0.99 | unsp | PcyM_0417100 | 84 S | QAPNSAESN | 0.992 | unsp |