

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
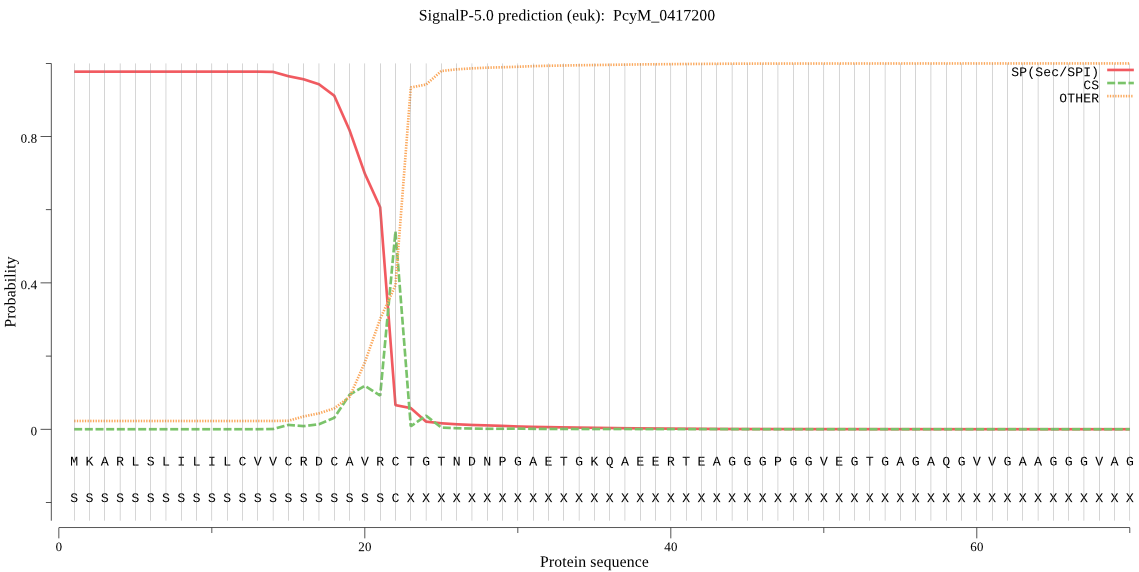
| PcyM_0417200 | SP | 0.009603 | 0.990346 | 0.000051 | CS pos: 22-23. VRC-TG. Pr: 0.7607 |
MKARLSLILILCVVCRDCAVRCTGTNDNPGAETGKQAEERTEAGGGPGGVEGTGAGAQGV VGAAGGGVAGTGAVPGQGGAVPGPGSGEAVSLSQDAGDTPREGQPVASQDGALPAENSED AQGRTDETSSNSVDSPSPDSNPNPVVDNAEAVTVTAPSEGPKEEIPVKSSLLKGYKGVKV TGPCSASFLVFFAPYLFIDVDAESSNIYLGTDLSDLEVTEKMGVGENNTNKCDDGKTFKF VALIGDDHLTIKWKVYDPKVQTPPQDDTAEMRKYVMKSLNGEFTSVQVHTVIQQNGSNVF ESKNYALSNDMPEQCDAIATNCFLSGSVYIEKCYRCTLKMKKLNPSDVCYNYIPKVENAP SQESISAQASDEESMQEKLAASIGMILQGMYKKGETGLNELLIFDEADAALKEELLNYCA LMKEVDTSGVLENYELGSEENVFANITNMLQKNSDYSVSSLQNKLKNPAMCLKNADEWVG SKMGLVLPNLPYNHLEVPHPSTPEVANVEDTSEDTQSGGYDGVIDFSTASKTNFSTSQYV DKMHCNGEYCDRTKDTSSCIAKIKAGDQGDCATSWLFASKVHLETIKCMKGHDHVASSAL YVANCSGKEAKDKCQTPSNPLDFLNTLEETKFLPAESDLPYSYKEVNNVCPEPKSHWQNL WENVKLLDKQYQPNSVSTKGYTAYQSDHFKGNMDAFNKLVKSEVMNKGSVIAYVKAQGVM TYDLNGKKVLSLCGGETPDLAVNIVGYGNYINGEGVKKSYWLLQNSWGKHWGDKGKFKVD MDGPPGCQHNFIHTAAVFNVDIPVVEKPTKEDAQIYNYYLKSSPDFWGNIYYKNVGGQTN TSAKNATGVANESVLHGQADTEVEVKVAGEDPATPLSTQATGGATAGQVSTVVPSLPVQV EVTVARNVGAPGDQAGSPGPAGPTGPAGSPGSPGQEGQEGPAGPPGTPGPEGPEGSAGPT GPEGPEGSAGPTGPPGPSGTPGTEGQPGTAPEAAVLDTQVSHVLKYIKKNKVKMNVVSYK NHEAITTGHDCWRSYSVNPDKYEECVKICEANWSKCENDVAPGFCLFEHGKDNDCFFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0417200 | 137 S | VDSPSPDSN | 0.992 | unsp | PcyM_0417200 | 137 S | VDSPSPDSN | 0.992 | unsp | PcyM_0417200 | 137 S | VDSPSPDSN | 0.992 | unsp | PcyM_0417200 | 370 S | SAQASDEES | 0.997 | unsp | PcyM_0417200 | 842 S | QTNTSAKNA | 0.991 | unsp | PcyM_0417200 | 1018 S | MNVVSYKNH | 0.992 | unsp | PcyM_0417200 | 99 T | DAGDTPREG | 0.996 | unsp | PcyM_0417200 | 135 S | NSVDSPSPD | 0.995 | unsp |