

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
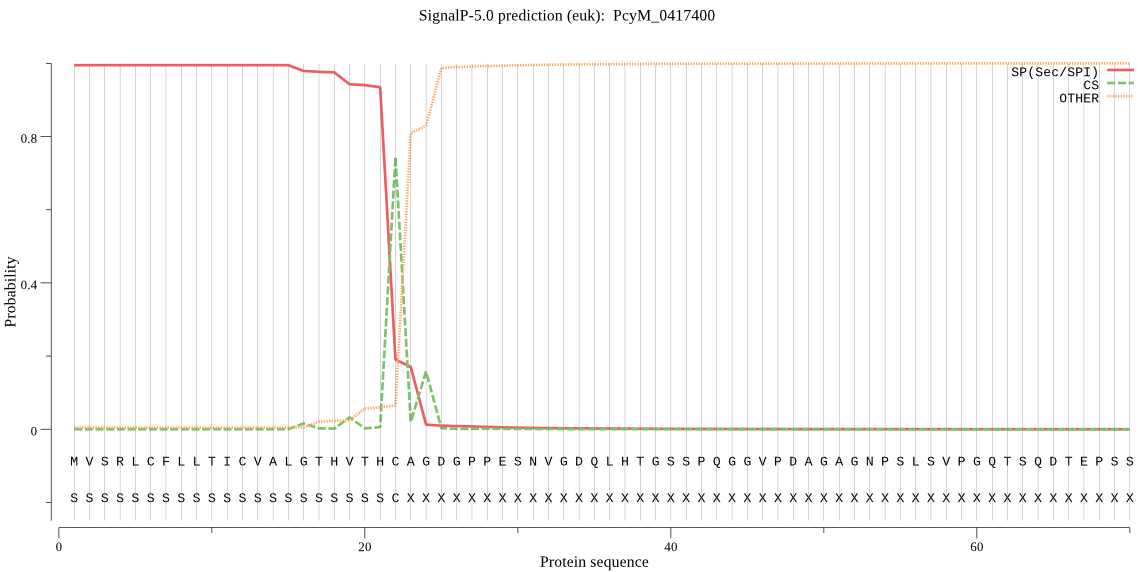
| PcyM_0417400 | SP | 0.000368 | 0.999631 | 0.000001 | CS pos: 22-23. THC-AG. Pr: 0.7863 |
MVSRLCFLLTICVALGTHVTHCAGDGPPESNVGDQLHTGSSPQGGVPDAGAGNPSLSVPG QTSQDTEPSSSSSENGIGSSSSNEETASPPTSADAKSNGIESESSDAASVQVEDTSEQNI QITSALLKHSNGIKITGSCKAHFRVFLSPHLWIYAVAAENKIQLRPNFGPITSIDLGNLI NDCKKGANNNFKFVVHTSDDILTVKWKVTGEGAAPGNQEDVKKYKLPPLDRPFTSVQVHS ANAKSKIIESKFYDIGSSMPAQCSVVATNCFLSGSLDIEHCYHCTLLEKKMAQNSECFKY VSKEAKELIEKDTPIKAQEEQAHSADHKLIESIDVILKAVYKSDKDDEKKELITLEEVDE NLKKELANYCTLLKEVDTSGTLNNHKLANEIETYRNLTRLLRMHSEENVVTLQDKLRNAA ICIKDIDKWIINKRGLSLPDGTPYYSEENTKEYLAEEFLKEIEKEKNACDDDAFDKDTNG VIDLNKIPGEMKFKSPYFKKSKYCNNEYCDRWKDKTSCISNIEVEEQGDCGLCWIFASKL HVETIRCMRGYGHFRSSALFVANCSKRSPEDRCNVGSNPTEFLQIVKDTGFLPLESDLPY NYSQAGNSCPNKRSKWTNLWGNTKLLYHKRPNHLFQTFGYVSYESKRFENNIDLFIDILK REVQNKGSVIIYIKTENVIDYDFNGKVVHSLCGHNDADHAANIIGYGNYINVHGEKRSYW LVRNSWGYYWGDEGNFKVDMYGPDGCKRNFIHTAVVFKIDLGIVEVPKKDDRPFYNYFIE YVPNFLYNLFYSSYDKGDDSNTLCENAKGGEAVGGDSVVDGQAVVTGQTETPTPGAAQDG AQPGAVGSEVNAAAGDNAGIVPPVGSQTGAVAQESGAVTPETGAVAQESGVVAPQAGAIA TQNGADTPQAGDIATQTGADTPQTGDIATQNGADTPQTGDIATQNGADTPQNGVPAPQTT AAASPPSVTPLNVTVEEGQRVGGVVSLIPVDNSGEKKIPVMHILKYVKETKMTRAFVRYD SLNEIESSHSCSRTYAKDEKGHEECIQFCLKNWASCRGHYSPGYCLTKMYAGSNCFFCSV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0417400 | 82 S | GSSSSNEET | 0.997 | unsp | PcyM_0417400 | 82 S | GSSSSNEET | 0.997 | unsp | PcyM_0417400 | 82 S | GSSSSNEET | 0.997 | unsp | PcyM_0417400 | 343 S | AVYKSDKDD | 0.998 | unsp | PcyM_0417400 | 437 S | KRGLSLPDG | 0.994 | unsp | PcyM_0417400 | 446 S | TPYYSEENT | 0.994 | unsp | PcyM_0417400 | 568 S | CSKRSPEDR | 0.997 | unsp | PcyM_0417400 | 1021 S | VRYDSLNEI | 0.994 | unsp | PcyM_0417400 | 1030 S | ESSHSCSRT | 0.995 | unsp | PcyM_0417400 | 71 S | EPSSSSSEN | 0.996 | unsp | PcyM_0417400 | 72 S | PSSSSSENG | 0.991 | unsp |