

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0510100 | OTHER | 0.999749 | 0.000193 | 0.000058 |
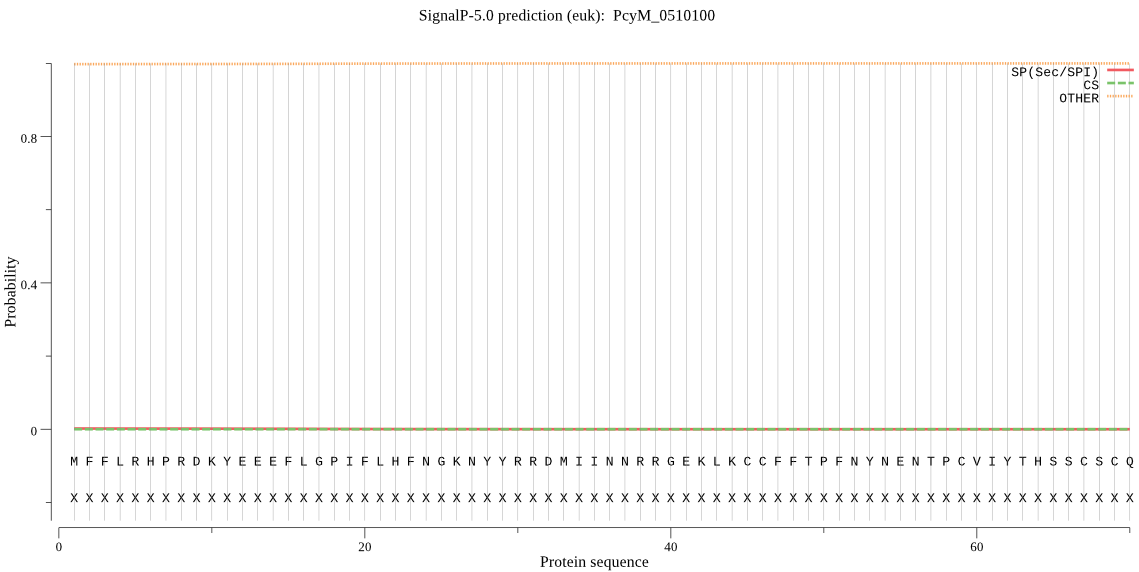
MFFLRHPRDKYEEEFLGPIFLHFNGKNYYRRDMIINNRRGEKLKCCFFTPFNYNENTPCV IYTHSSCSCQLEALDILHILLICECSIFSYDCSGCGLSDGYFSTSGWNESQDLFLILNHL RNVEHVKNFALWGKYSGAVSSIIVASLDANIKLLIVDSPYVSLTELYKTTFHLSAKGKAE IIFKNICLYFAKRKIKKKFHYDIENICPVFFIEDITIPTIYIISRNDKIVHPAHTLYLAY KQKSQQKIIYTCEKAAHAYESFSYDNKLTAAIRTVLFGAEKGEVQNMFSVYTYMNVFNGL MEKYAHEFCFIDSLIQKKLGKKNKIIDKVKRFICFKYQSKASLTSSSCFNQSTIDSIRDA STTGENDYMQYEPSKNGLCWKFKSVSGEQDERDNHGLSEEQDPREDHGLADEYEYPFRDV NDGCDVNDGCDDHNFMGGKDATENLLDASPLKGYDANKVPAFRSTFHAGEQSGQRRFNLM HMQSARSSLKSKKNTYKKSLTWDTKLQSAVTYSKEDCPFELLKS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0510100 | 398 S | NHGLSEEQD | 0.992 | unsp | PcyM_0510100 | 398 S | NHGLSEEQD | 0.992 | unsp | PcyM_0510100 | 398 S | NHGLSEEQD | 0.992 | unsp | PcyM_0510100 | 488 S | SARSSLKSK | 0.995 | unsp | PcyM_0510100 | 491 S | SSLKSKKNT | 0.997 | unsp | PcyM_0510100 | 513 S | AVTYSKEDC | 0.991 | unsp | PcyM_0510100 | 356 S | STIDSIRDA | 0.992 | unsp | PcyM_0510100 | 386 S | FKSVSGEQD | 0.993 | unsp |