

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
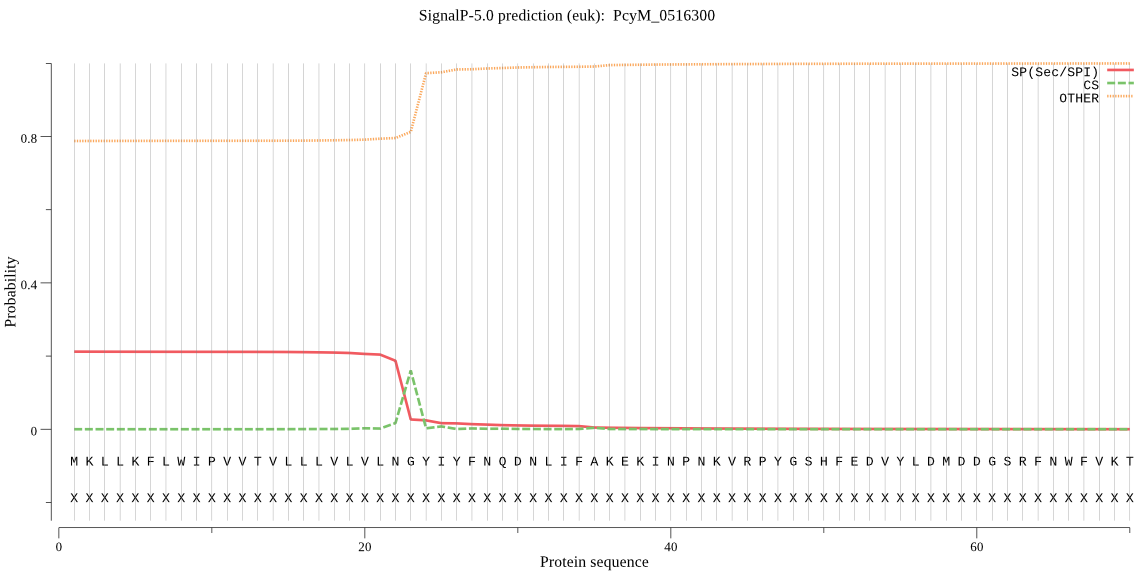
| PcyM_0516300 | SP | 0.432468 | 0.565862 | 0.001671 | CS pos: 23-24. LNG-YI. Pr: 0.8329 |
MKLLKFLWIPVVTVLLLVLVLNGYIYFNQDNLIFAKEKINPNKVRPYGSHFEDVYLDMDD GSRFNWFVKTKGHENKPVFLYYLGKGGYIEKYVKLFDMIVQRVDVSIFSCSNRGCGTNEG TPSEEQFYKDALVPLNYLKQKHTKQLFIFGNSMGGAVALETASRHQKDIYGVIVENPFLS IKKMSEQNYPFLNFFLLSFEILIRSKMDNTEKIKRIHVPLLINTSERDEMVPPEHSRILF ELCPSKYKFRYISKKGTHNNIIKVDDGSYHASLKKFVQTAINIRERNAEPQAVPPLPAAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0516300 | 55 Y | FEDVYLDMD | 0.991 | unsp | PcyM_0516300 | 253 S | FRYISKKGT | 0.996 | unsp |