

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
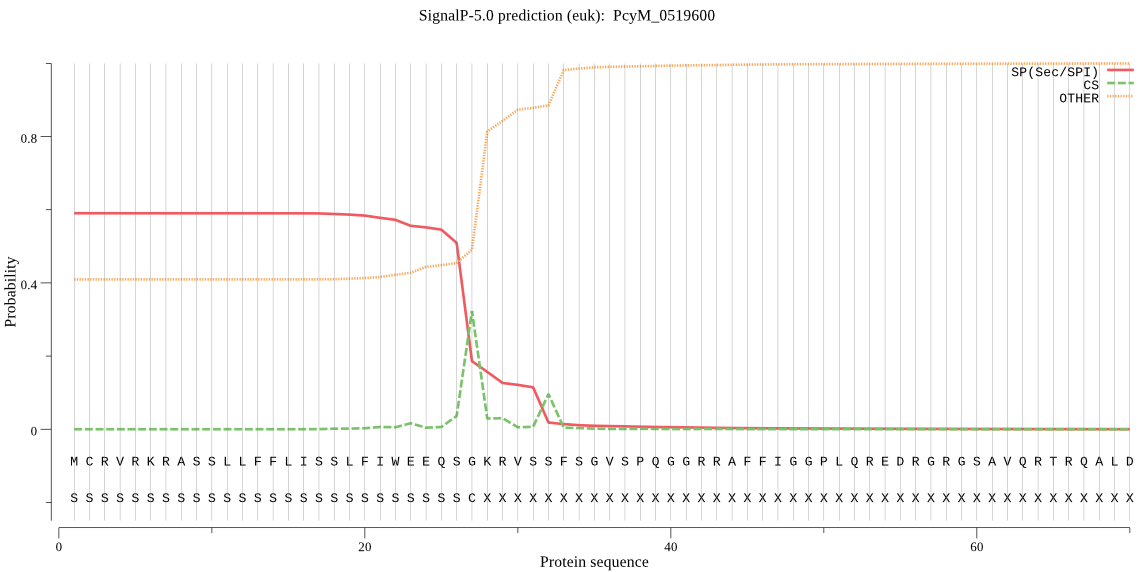
| PcyM_0519600 | SP | 0.059451 | 0.938372 | 0.002177 | CS pos: 27-28. QSG-KR. Pr: 0.8073 |
MCRVRKRASSLLFFLISSLFIWEEQSGKRVSSFSGVSPQGGRRAFFIGGPLQREDRGRGS AVQRTRQALDGVQKKHLHADEAVGSSRRGAANGRLFYSVGSHPGEKNFENGGTYERRGEH GSAHLSAHRSVHQSAHRSERAFGRTAALRMSDEEYTINSDDYTEKAWEAITTLNKIGEKY ESAYVEAEMLLLALLNDGPEGLAQRILKESGIDTDLLMQEIDEYLKKQPKMPSGFGEQKI LGRTLQAVLSTSKRLKKEFHDEYISIEHLLLGIIAEDSKFTRPWLLRYNVNYEKVKKATE RVRGKKKVTSKTPEMTYQALEKYSRDLTALARAGKLDPVIGRDTEIRRAIQILSRRTKNN PILLGDPGVGKTAIVEGLAIKIVQGDVPDSLKGRKLVSLDLSSLIAGAKYRGDFEERLKS ILKEVQDAEGQVVMFIDEIHTVVGAGAVAEGALDAGNILKPMLARGELRCIGATTVSEYR QFIEKDKALERRFQQILVEQPSVDEAISILRGLKERYEVHHGVRILDSALIQAAVLSDRY ISYRFLPDKAIDLIDEAASNLKIQLSSKPIQLDNIEKQLVQLEMEKISILGDSPRGGAVS RGSSGTAGSTAISSAISSGAGGGPAQGEETPIDYSQSPNFLKKKINEKEINRLKMIDHIM SQLRKEQKRILESWSTEKGYVDNIRAIKERIDVVKVEIEKAERYFDLNRAAELRFETLPD LENQLKKAEGNYLNDIPEKNRMLKDEVTSEDIMNIISLSTGIRLNKLQKSEKEKILNLEN ELHKQIIGQDDAVRIVSKAVQRSRVGMNDPKRPIASLMFLGPTGVGKTELSKVLADVLFD TPDAVIHFDMSEYMEKHSISKLIGAAPGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHP DVYNLLLRVIEEGKLTDTKGNLANFRNTIIIFTSNLGSQSILDLANDPNKKEKIKEQVMK SVRETFRPEFYNRIDDHVIFDSLSKKELKQIANIEIEKVANRLIDKNFKISIDDAVFSYI VDKAYDPAFGARPLKRVIQSEIETEIAIRILNETFVENDTIRVSLKDGALHFSKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0519600 | 390 S | DVPDSLKGR | 0.991 | unsp | PcyM_0519600 | 390 S | DVPDSLKGR | 0.991 | unsp | PcyM_0519600 | 390 S | DVPDSLKGR | 0.991 | unsp | PcyM_0519600 | 542 S | DRYISYRFL | 0.993 | unsp | PcyM_0519600 | 593 S | ILGDSPRGG | 0.993 | unsp | PcyM_0519600 | 770 S | KLQKSEKEK | 0.996 | unsp | PcyM_0519600 | 961 S | QVMKSVRET | 0.997 | unsp | PcyM_0519600 | 984 S | FDSLSKKEL | 0.997 | unsp | PcyM_0519600 | 1011 S | NFKISIDDA | 0.993 | unsp | PcyM_0519600 | 1040 S | RVIQSEIET | 0.991 | unsp | PcyM_0519600 | 1064 S | TIRVSLKDG | 0.998 | unsp | PcyM_0519600 | 151 S | ALRMSDEEY | 0.997 | unsp | PcyM_0519600 | 210 S | ILKESGIDT | 0.997 | unsp |