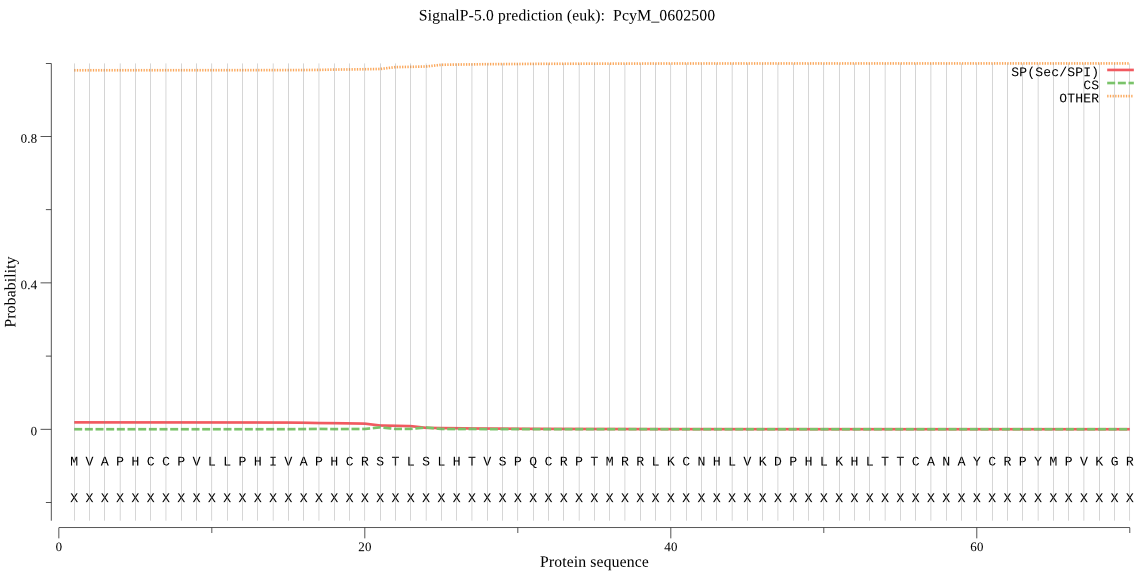
Fasta :-
MVAPHCCPVLLPHIVAPHCRSTLSLHTVSPQCRPTMRRLKCNHLVKDPHLKHLTTCANAY
CRPYMPVKGRPTWSTDRKRIRTSIFKDDDIYMDPLQIHNEEPNKTNYLNRASPDECVYTR
NNVGMNAVLINKKYMDLKCINQLYKELLNGEINFTKRFTFITSISNESFNYGFNLLDMLK
IVEVYQKGKNNKHADVLKKILYNINELSYLIFSYRKPLIVYCNGSVRGSGGFIPFLANNS
ASYFHSSYAYTNLKYSFLPYGGISYVLANLRGSIGFYLALTGQVIQSADLMWCGLTKRWL
AEDCLELMELTSESQLEVSEQDANLLLEEHFLKVPKIYTLKCYEEIIHDHFKYPSLMQIM
AKLDASRKRTHSDERVRLWAEKTYQQICSQPPIAAHLTFEILHLLRTHKMDLLKKAQVTK
KLYNQMIQNSYKVVPTTKEEVSLAELKLTIDSELLIKALNIETNAIANFISCPDLLNGIT
SYLVKDTDHSFKCTYLNNSLLETKKDIIHYFLFYKNDYEFTVHDRPDISFSSLSVLDRCN
QPYGSRDRHFYAEQSKRWSDDYLKDQLNEINKLAL