

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
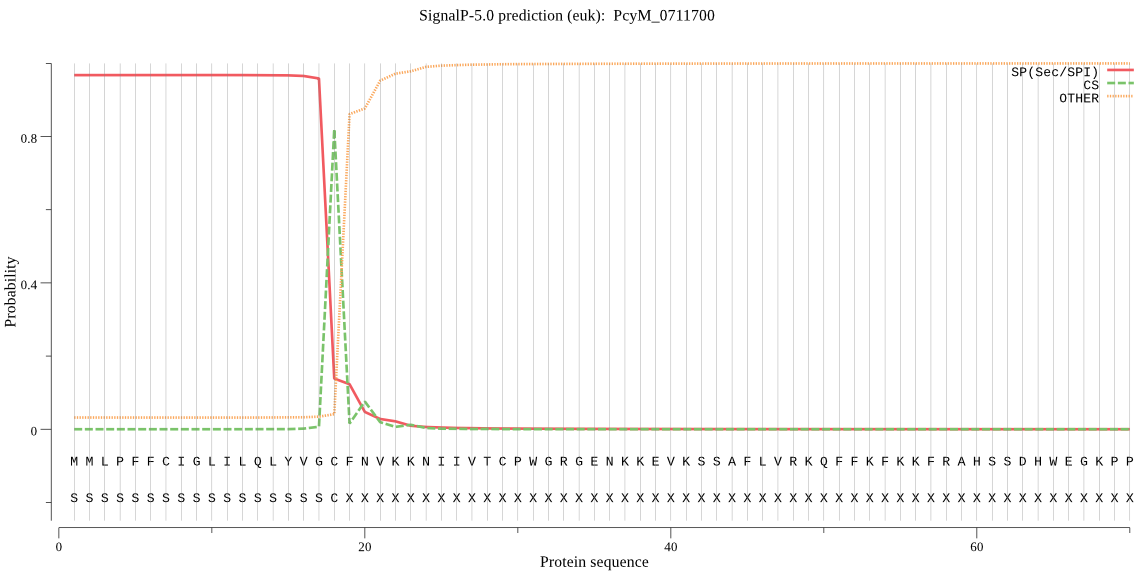
| PcyM_0711700 | SP | 0.286592 | 0.706281 | 0.007126 | CS pos: 18-19. VGC-FN. Pr: 0.6167 |
MMLPFFCIGLILQLYVGCFNVKKNIIVTCPWGRGENKKEVKSSAFLVRKQFFKFKKFRAH SSDHWEGKPPIDAAQNGAKNTLSEFYHRNCRRLRKTFELVKSGLISIQDNLFSNSLASQL AISVSFFLLHFYMLSRHFLILFPYQLIPNHSNILMSLDMNNTLFFLMSIFFFKRFKAHFH SFKQRLQLNRFTIKLQDHKRKNILSVCFFLIASYVLSGYVSIYTERILTLWKLFSHPRSD NVVKSLQILTGHFIWVACSIVVFKQLLYPYFLNNESNLNLRHKDSWYFEVIYGYVFSHFV FSIVDLLNNFIINRFMGEAQDEVYVDNSIDDIVSGREFVSTLLCIISPCFSAPFFEEFIY RFFVLKSLNLFMHIHYAVTFSSLFFAIHHLNIFNLLPLFFLSFLWSYIYIYTDNILVTML IHSFWNIYVFLSSLYS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0711700 | 334 S | DDIVSGREF | 0.991 | unsp |