

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0716600 | OTHER | 0.993163 | 0.000413 | 0.006425 |
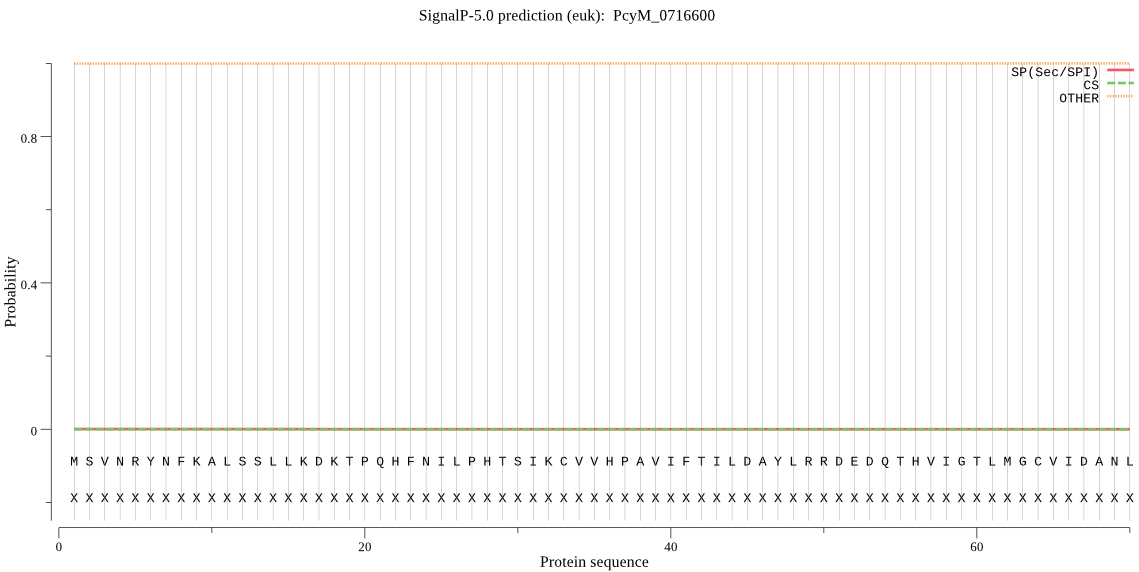
MSVNRYNFKALSSLLKDKTPQHFNILPHTSIKCVVHPAVIFTILDAYLRRDEDQTHVIGT LMGCVIDANLIEISDCFVDKHSLNEGGFLQIIKDHHETMYELKQKIRPRDQVVGWFCSGS ELSELSCAVHGWFKEHNSISKFYPHSPLNEPIHLLVDAALESGFLNIKAYVQLPITLVKE YFVHFHEIQAELLPSNVERAEVLQYQEKGLTGKDKDSHMHGNKNSVLPNDMNEMSLKKLL IMLKQCKSYVQDVIDKKKKGNLEVGRYLHKVFSNDSFSTLEKFDSINESILQDNLMISYL SNLAHLQFLIAEKLNASSLQ