

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0824100 | OTHER | 0.999985 | 0.000001 | 0.000014 |
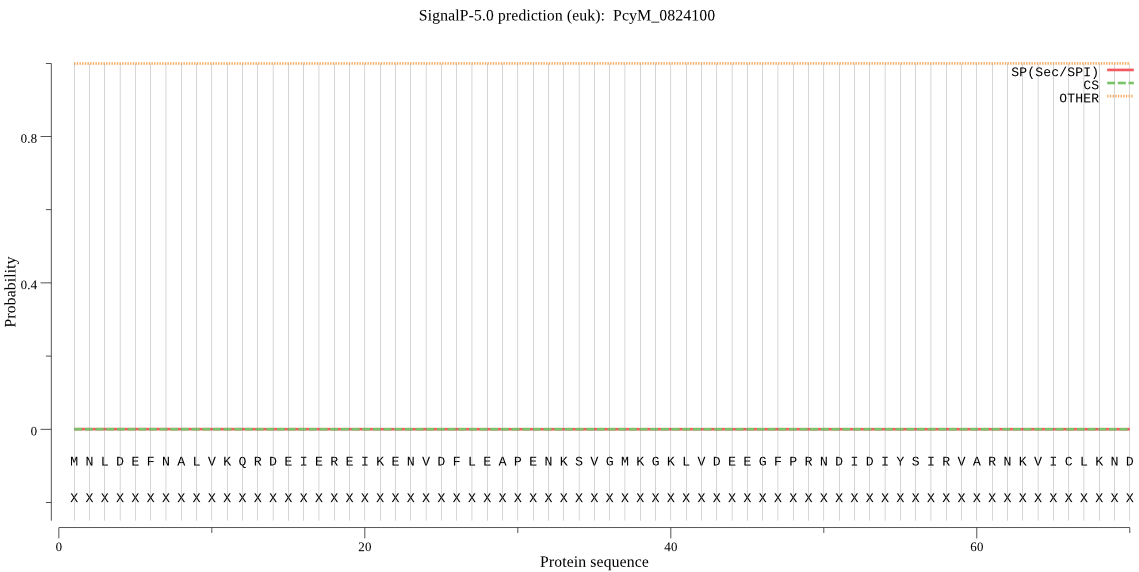
MNLDEFNALVKQRDEIEREIKENVDFLEAPENKSVGMKGKLVDEEGFPRNDIDIYSIRVA RNKVICLKNDYLNVNKKIEEYLHKVHNSHPVIRVQRSKAKDEQGNDPNESSPESVTEDYD ESAPDYEKLIEEARRSTFAMIDEMVENSPSHKAGLRINDYIIQFGDIRKKKSEKKKSDKN EKENDNERESEKDHEDIFKRIAAYMSNNPTRIKVKILREGKIFFYFVFPSKTANGLYIGC HLTPTRGVVV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0824100 | 177 S | EKKKSDKNE | 0.997 | unsp | PcyM_0824100 | 177 S | EKKKSDKNE | 0.997 | unsp | PcyM_0824100 | 177 S | EKKKSDKNE | 0.997 | unsp | PcyM_0824100 | 190 S | NERESEKDH | 0.998 | unsp | PcyM_0824100 | 114 S | SSPESVTED | 0.996 | unsp | PcyM_0824100 | 172 S | RKKKSEKKK | 0.996 | unsp |