

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0827900 | SP | 0.075413 | 0.669823 | 0.254764 | CS pos: 23-24. QHA-RV. Pr: 0.9579 |
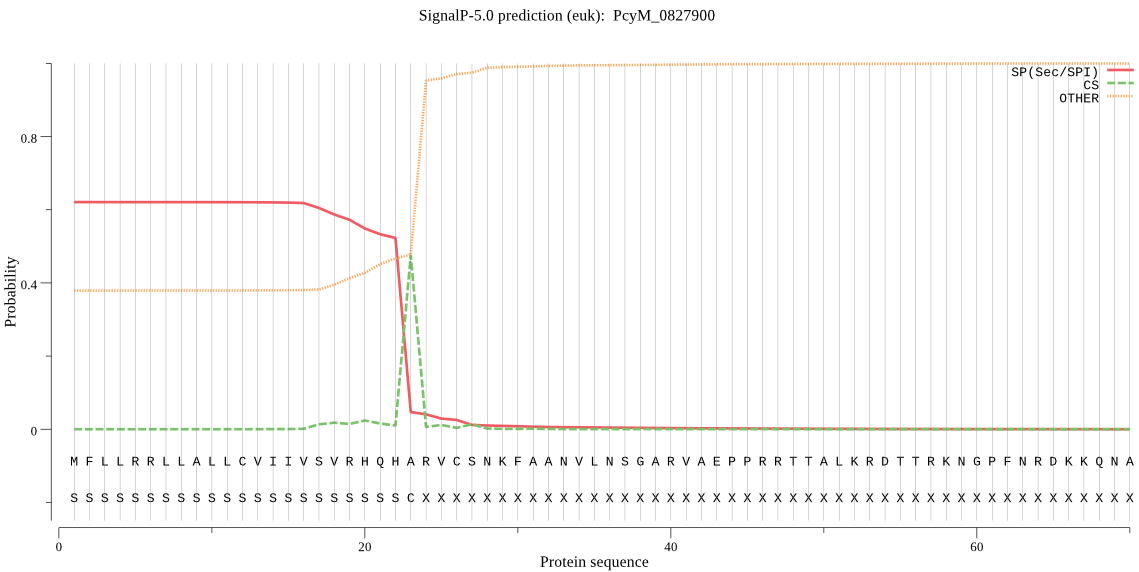
MFLLRRLLALLCVIIVSVRHQHARVCSNKFAANVLNSGARVAEPPRRTTALKRDTTRKNG PFNRDKKQNATSQHGYYFTKEKNKIVKKKKKFSLNYSRTPKTAAGGRRTKARTHYPLLFI SPSSTSLNEITESVKALFNLDYIENSYLYSYIKSFKRTPPLTKIYLLCTLFLSICMHANK NIYKLIVFDFGKIFYEGQAWRLITPYFYIGNLYLQYFLMFNYLHIYMSSVEIAHYKNPED LLTFLTFGYLSNLLFTIIGSMYNENVLNLQWYVNKLWRVILLGGGKSTSTKADTDVRIAK EQYNHLGYVFSTYILYYWSRINEGTLINCFELFLIKAEYVPFFFIIQNVLLYNEFSLFEV ASILSSYFFFTYEKNLQLNFVRRFNLALLKALRVYPMYEAYREEYE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0827900 | 289 S | GKSTSTKAD | 0.992 | unsp |