

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
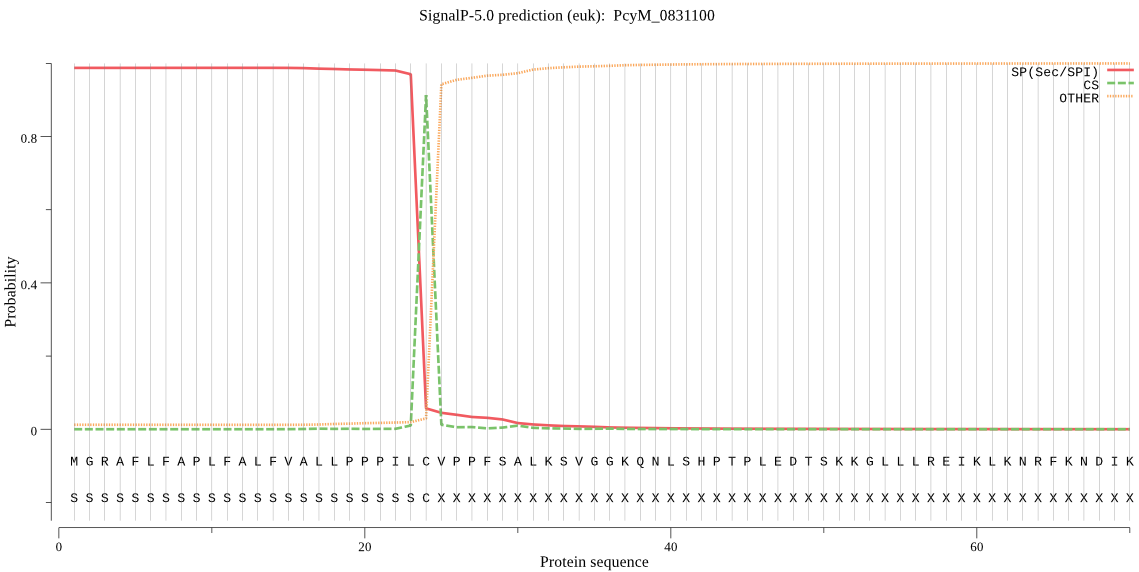
| PcyM_0831100 | SP | 0.001652 | 0.998089 | 0.000259 | CS pos: 24-25. ILC-VP. Pr: 0.8972 |
MGRAFLFAPLFALFVALLPPPILCVPPFSALKSVGGKQNLSHPTPLEDTSKKGLLLREIK LKNRFKNDIKGFIKNVRSFHDIIEDRTPNSLLYVQEDLLNFHNSQFIGDIEIGTPPQSFK VVFDTGSSNFALPSTKCVKGGCVSHKKFNPDQSRTYTRQLKGNKESIYTYIQYGTGKSIL EHGYDDVNLKGLRIKHQSVGLAIEESLHPFSDLPFDGIVGLGFSDPDFSFQKGYGTSLIE TIKKQNLLKRNIFSFYVPKNLKEPGSITFGRANSKYALEGRKIEWFPVISIYFWEINLID VQLSDNNLNMCENRKCRAAIDTGSSLLTGPSSLMQPLIEKLYLQKDCSNKSSLPNISFIL KNVEGKEVKFDFTPDDYILEETDEEDNSVQCVIGIMSLDVPAPRGPIFVFGNVFIRKYYS IFDNDHKLVGLTEARHDW
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0831100 | 420 S | RKYYSIFDN | 0.995 | unsp |