

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0836800 | SP | 0.160107 | 0.824749 | 0.015144 | CS pos: 20-21. THA-KR. Pr: 0.9742 |
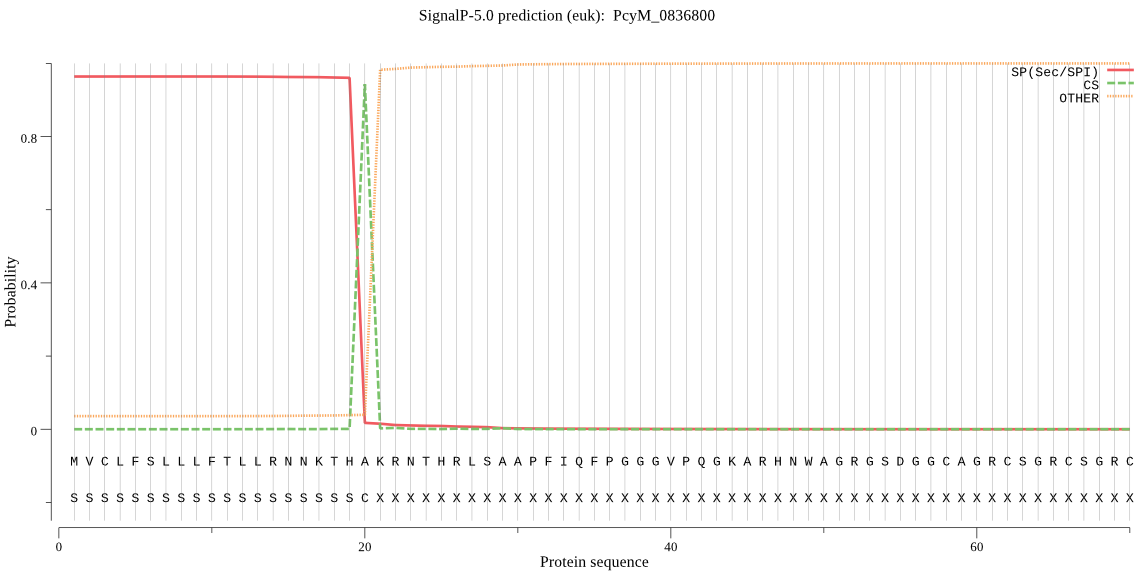
MVCLFSLLLFTLLRNNKTHAKRNTHRLSAAPFIQFPGGGVPQGKARHNWAGRGSDGGCAG RCSGRCSGRCSGVYGRCSKTRGASSNSLPLSIGDEQEGVEPGEESASEGAADGVSRSHPP AKGMKIVKNGELCMDPATGVGGFLVGEGTRLEDPPPKGKQIHKSSRGGSGIISRRSGIIS SSSSRRYRRREGLLPRVDHIKDMKKDVKLFFFKKRIIYLTEEINKKTADEMISQLLYLDN INHDDIKIYINSPGGSINEGLAILDIFNYIKSDIQTISFGLVASMASVILASGKKGKRKS LPNCRIMIHQPLGNAFGHPQDIEIQTKEILYLKKLLYNYLSTFTNQTTETIEKDSDRDYY MNALEAKRYGIIDEVIQTKLPHPYFDEAVRESSSGAPSV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0836800 | 355 S | IEKDSDRDY | 0.998 | unsp | PcyM_0836800 | 355 S | IEKDSDRDY | 0.998 | unsp | PcyM_0836800 | 355 S | IEKDSDRDY | 0.998 | unsp | PcyM_0836800 | 392 S | AVRESSSGA | 0.991 | unsp | PcyM_0836800 | 176 S | ISRRSGIIS | 0.99 | unsp | PcyM_0836800 | 183 S | ISSSSSRRY | 0.995 | unsp |