

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
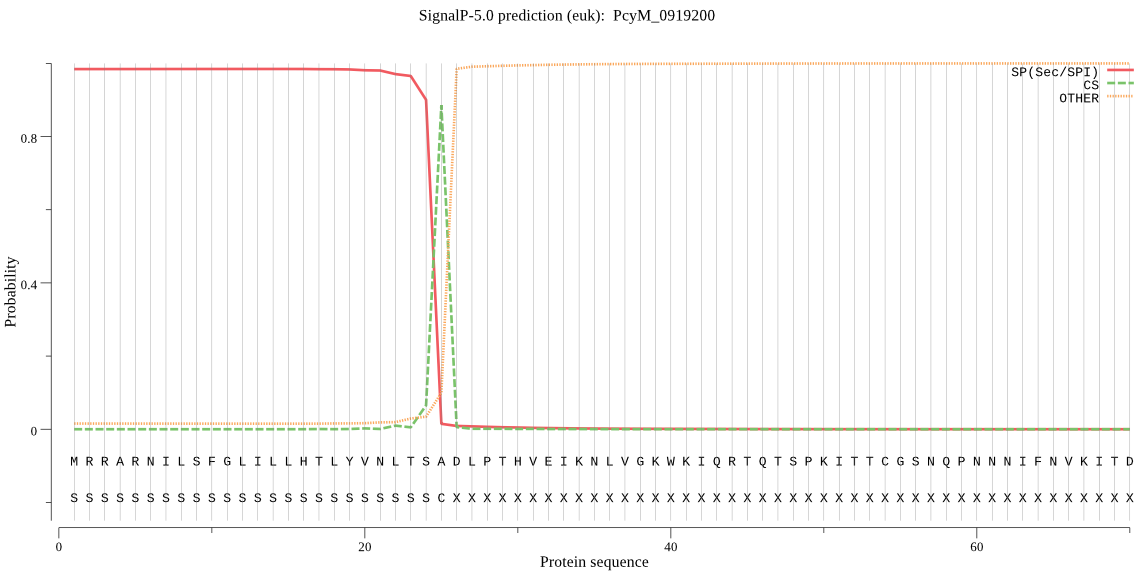
| PcyM_0919200 | SP | 0.001201 | 0.998726 | 0.000073 | CS pos: 25-26. TSA-DL. Pr: 0.9368 |
MRRARNILSFGLILLHTLYVNLTSADLPTHVEIKNLVGKWKIQRTQTSPKITTCGSNQPN NNIFNVKITDYKKYLLDNHYKFTSELYVILSDDFVPYGDMHDTTGNEHRKNWKVLVVYDE HKRKIGTWTTICDEGFEIRLGNETYTAFMHYEPTGKCGEAKEDDATDTNGETDCYVTNFN KIRFGWVDISNSNNEKLYGCFYAERYRVSGESDNASNTQLNFANGTNINSVLKMHNFLHT TPAATTSSSNDKPTFTKRKNMHIDQNSELYWHKMKHHGKKKPITKAMILNSKQKYACPCN KDEHVQNDVNNGGTPDQPVSPVSLMQLGSGSGEDETNEMDLENYEDTEKSPHRELEIDEL PKNFTWGDPFNKNTREYDVTNQLLCGSCYIASQMYVFKRRIEIGLTKNLDKKYLNDFDDL LSIQTVLSCSFYDQGCNGGYPYLVAKMAKLQGIPLDKVFPYTATTETCPYPVDQSGVTSL SGSNSSNTSTNNLRQINAVMFGTKTTNDMHDHFESNISDDPDKWYAKDYNYIGGCYGCNQ CNGEKIMMNEIYRNGPIVASFEATPDFYDYADGVYYVKDFPHARRCTVDDTKSNFVYNVT GWEKVNHAIVLVGWGEEEIDGTMYKYWIGRNSWGKAWGKEGYFKIIRGVNFSGIESQTLF IEPDFTRGAGKILLEKLRNE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0919200 | 331 S | LGSGSGEDE | 0.994 | unsp |