

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0919300 | SP | 0.045589 | 0.951581 | 0.002830 | CS pos: 27-28. ASC-SA. Pr: 0.4987 |
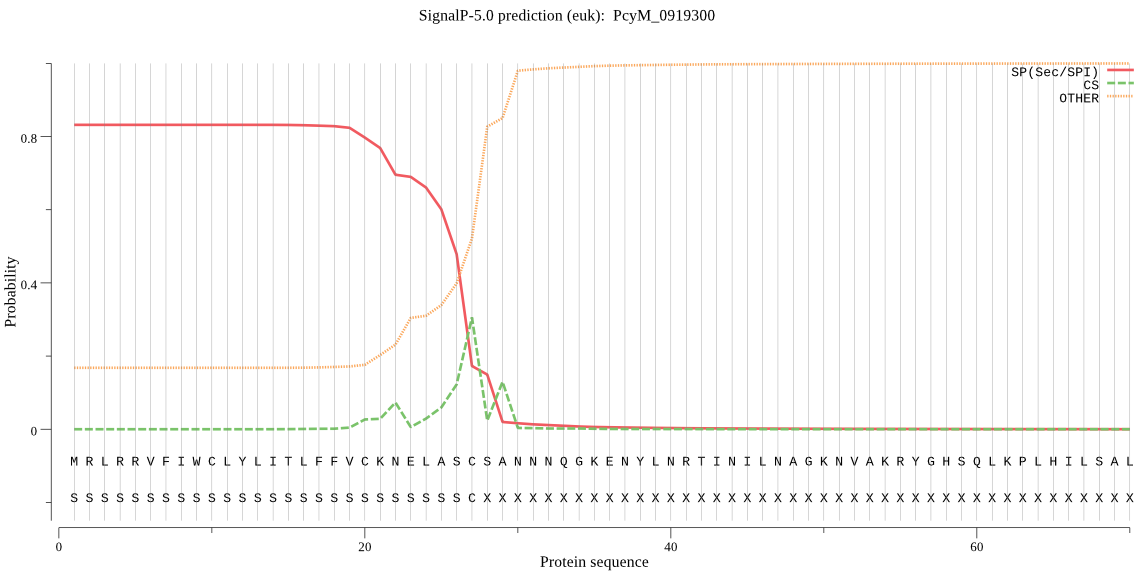
MRLRRVFIWCLYLITLFFVCKNELASCSANNNQGKENYLNRTINILNAGKNVAKRYGHSQ LKPLHILSALAKSDYGSNLLKENSVNASNLKEYIDTALEQTRAGAPLDNKSKIGYSDEVK EVLAEAEALANKYKSQKVDVEHLLSGLMNDDLVNEIMNEVYLTEEAVKGILKNKLEKNKK DKDGKSGGLYLEQFGSNLNEKVRNGKLQGIYGRDEEIRAVIESLLRYNKNSPVLVGQPGT GKTTIVEGLVYRIEKGDVPKELRGYTVISLNFRKFTSGTSYRGEFETRMKNIIKELKNKK NKIILFVDEIHLLLGAGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFE RRFEKILVEPPSVENTIKILRSLKSKYENFYGIHITDKALVAAAKISDRFIKDRYLPDKA IDLLNKACSFLQVQLSGKPRIIDVTEREIERLAYEISTLEMDVDKVSKRKYNNLIKDFEN KKEQLKKHYEEYVISGERLKRKKETEKKLNELKELAQNYISANKEPPIELQNSLKEAQEK YMEVYKETLAYVEAKTHNAMNVDAVYQEHVSYIYLRDSGMPLGSLSFESSKGALKLYNSL SKSIIGNEDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFSSK DNLIRVNMSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVRERPHSVVLFDELEKAHPDV FKVLLQILGDGYINDNHRRNIDFSNTIIIMTSNLGAELFKKKLFFDANNSDTPEYKRVFD DLRIQLIKKCKKVFKPEFVNRIDKIGIFEPLSKKNLREIVKLRFKKLEKRLEEKNIHVSV SEKAIDYIIDQSYDPELGARPTLIFIESVIMTKFAIMYLKKELVDDMDVHVDFNKAANNL VINLTAV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0919300 | 372 S | VEPPSVENT | 0.991 | unsp | PcyM_0919300 | 372 S | VEPPSVENT | 0.991 | unsp | PcyM_0919300 | 372 S | VEPPSVENT | 0.991 | unsp | PcyM_0919300 | 533 S | ELQNSLKEA | 0.995 | unsp | PcyM_0919300 | 658 S | IELFSSKDN | 0.996 | unsp | PcyM_0919300 | 706 S | ERPHSVVLF | 0.994 | unsp | PcyM_0919300 | 812 S | FEPLSKKNL | 0.994 | unsp | PcyM_0919300 | 841 S | HVSVSEKAI | 0.995 | unsp | PcyM_0919300 | 135 S | NKYKSQKVD | 0.995 | unsp | PcyM_0919300 | 280 S | TSGTSYRGE | 0.992 | unsp |