

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_0931000 | OTHER | 0.998180 | 0.000362 | 0.001458 |
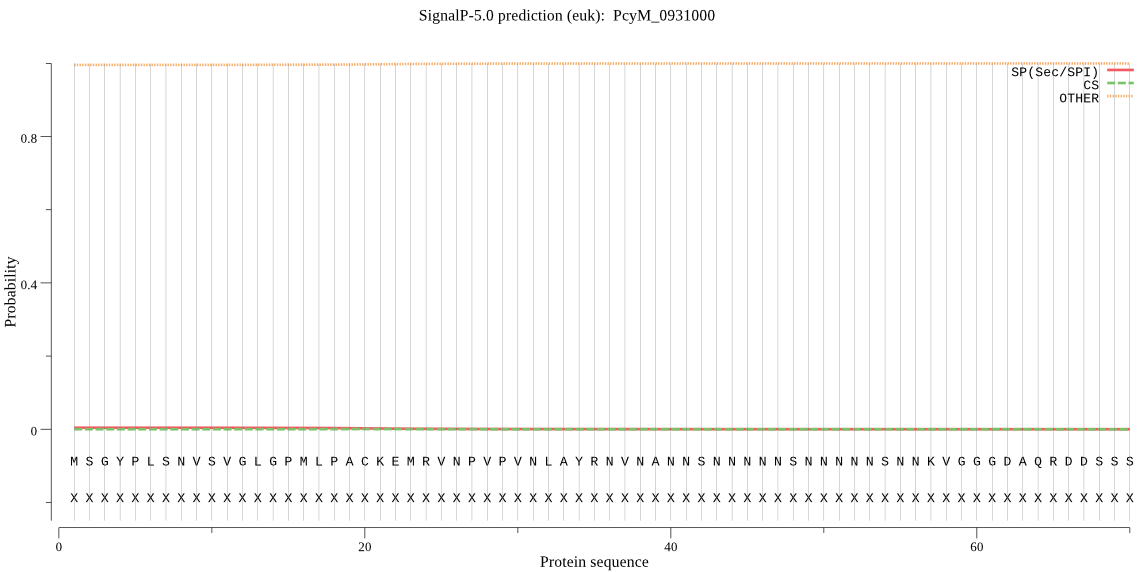
MSGYPLSNVSVGLGPMLPACKEMRVNPVPVNLAYRNVNANNSNNNNNSNNNNNSNNKVGG GDAQRDDSSSSPQRIDTRSSAKQSNNKSDVKDEKKTITECTPYKFGKRKKKNNNNNLDSM EELLFKLTNGKFKAEKHHVYTSDGYRLNLYRIVSTNKKENLSKKKQVFCLNHGLFESSIS YTCKGYESLAFQIFANNYDVWVSNNRGNAFTKFVGKNYAMKKLRERYSLQDLMDIGVDVS EEMAVQGSSSATGDEEKKCTESGSSSGEESCSGEESSRGGESHLGEERKTTNLRKAGNAS DNTTNDNGTKNSEGNNKSEDESRTVAPSCGSPGANLSIGDAPSGEGGSKVGTNAPSDGNN KAEKQTQCSGSSSANEGTEDAPSNVGTRSFMDIFKRIKGIKGTDKNEKNCKKVYCDIDVE TADEMDPTVSADDSTTVNGEEEEQQHQKQEHKQLKKKEDEDENGVSAHETGDSQNPPGPF GDASNCDSNTGCSFPHVPKEVENGEVEGGNAAEGEAQGEDEEEVEELVNLNKQSEVEEEG EYESEYESEYESEHEGDEDDAIELDEPEIDNWTFEDMGTKDLPVVIKYIRKKTQRDKIVY VGFSQGSVQLLVSCCLNDYVNSSIKRTYLLSLPIILKSKDELLKSVKLLLIASKWYKAII GSKEFLQNVLPEKMSTNLICNSADLFTRNFFKFYTNDLDDNYKKIYYKHTPSGSTSKANL KKWKSSLHDGPVSEAIDKYAYKCSFPITLVYGIKDCLVDAVRSIEYMKKKFTKNDLKIIT DPEWSHIDPVLTDHRNVVLSCILEDMKRDEEEERKKQEEEKQEGEKQEGEKQEGEKQEEE KQEEEKQEVEKQEEEKQEGEKEQTQEEEKEQTQEENKQE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0931000 | 80 S | DTRSSAKQS | 0.996 | unsp | PcyM_0931000 | 80 S | DTRSSAKQS | 0.996 | unsp | PcyM_0931000 | 80 S | DTRSSAKQS | 0.996 | unsp | PcyM_0931000 | 162 S | KENLSKKKQ | 0.996 | unsp | PcyM_0931000 | 265 S | ESGSSSGEE | 0.996 | unsp | PcyM_0931000 | 266 S | SGSSSGEES | 0.998 | unsp | PcyM_0931000 | 272 S | EESCSGEES | 0.995 | unsp | PcyM_0931000 | 544 S | GEYESEYES | 0.994 | unsp | PcyM_0931000 | 548 S | SEYESEYES | 0.997 | unsp | PcyM_0931000 | 552 S | SEYESEHEG | 0.996 | unsp | PcyM_0931000 | 623 S | YVNSSIKRT | 0.995 | unsp | PcyM_0931000 | 726 S | KWKSSLHDG | 0.995 | unsp | PcyM_0931000 | 785 S | DPEWSHIDP | 0.993 | unsp | PcyM_0931000 | 68 S | QRDDSSSSP | 0.992 | unsp | PcyM_0931000 | 71 S | DSSSSPQRI | 0.995 | unsp |