

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
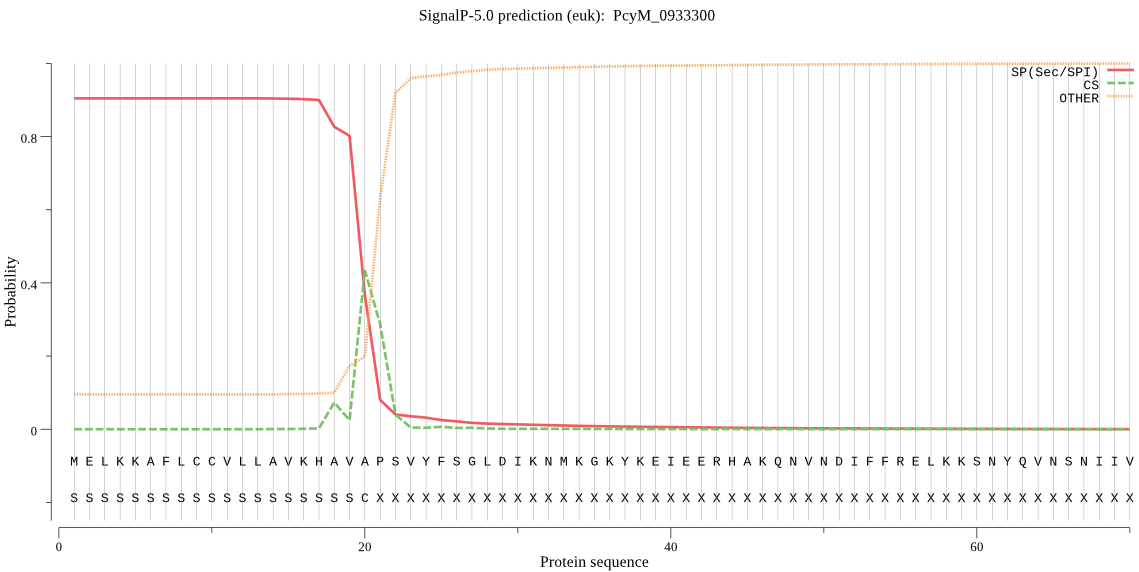
| PcyM_0933300 | SP | 0.025495 | 0.972400 | 0.002104 | CS pos: 18-19. KHA-VA. Pr: 0.3353 |
MELKKAFLCCVLLAVKHAVAPSVYFSGLDIKNMKGKYKEIEERHAKQNVNDIFFRELKKS NYQVNSNIIVLSTSRHYFNYRHTSNLLTAYKYLKHVGDNMDRNILLMVPFDQACNCRNIV EGTIFNEYEKPPSEDLKKKKMKENLYSHLNIDYKNDNIRDEQIRRVIRHRYDALTPAKYR LYTNGNREKNLFIYMTGHGGVSFFKIQDFNIVSSAEFSLYIQELLIKNIYKYIFVIIDTC QGYSFYDKVLDFLKKNKINNVFLMSSSDKNENSYSLHSSKYLSVSTVDRFTFYFFSYLEN MNRIYTHEPHKNAKAFSLYTILNYLKTQHLISTPTINNSKFSASMFMHSKNILFYDSSGF YVSKDAEESALDGLGKHSYNVSQKADKTAIRSSCLGDLSACGHMKNEIYTHMGNLYSRSS FYNNVEVYFKEESHFTDYYFTSERFAGGYLLPAFLFLVVIALCLLFFLFT
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_0933300 | 420 S | YSRSSFYNN | 0.992 | unsp |