

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
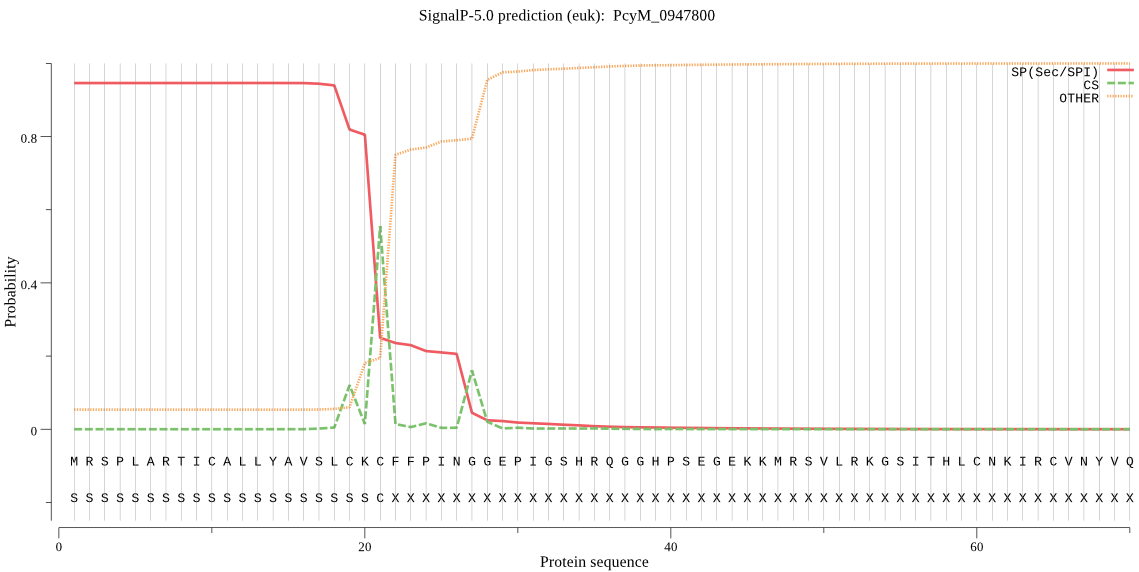
| PcyM_0947800 | SP | 0.016871 | 0.980519 | 0.002610 | CS pos: 21-22. CKC-FF. Pr: 0.6202 |
MRSPLARTICALLYAVSLCKCFFPINGGEPIGSHRQGGHPSEGEKKMRSVLRKGSITHLC NKIRCVNYVQWYEGRRRISSATWNSGKRSPVSSRLFGSFETKIPGSQADLVVDGIAVSIS DNTNIFKEETSSTPIVLIHGCYGSKNNFRVFSKSLKSSKIVTLDLRNHGNSKHTDSMKYE EMESDIKKVLDELHIRKCCLVGFSLGGKVSMYCALKNQSLFSHLVVMDILPFDYNEKKCH VKLPYNISYMTKILFNIKTKMRPRSKAEFVAYLRAEVPDISSSFEQFICTSLREEVAKVG KAAKEANEAKAEKAAKEADAAGNAAGVMDKPMGGTDFVGAADAIGCSEGPAAKQQKNLVW KINVDTIWRELPHILSFPLNHQERKYHNPCSFIIGKKSDLVYTMPQYQTIIKNYFPSSEN FILPDATHTVYIDNAKECADIVNRTLRL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_0947800 | 176 S | KHTDSMKYE | 0.993 | unsp | PcyM_0947800 | 176 S | KHTDSMKYE | 0.993 | unsp | PcyM_0947800 | 176 S | KHTDSMKYE | 0.993 | unsp | PcyM_0947800 | 265 S | MRPRSKAEF | 0.996 | unsp | PcyM_0947800 | 106 S | KIPGSQADL | 0.993 | unsp | PcyM_0947800 | 154 S | VFSKSLKSS | 0.996 | unsp |