

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
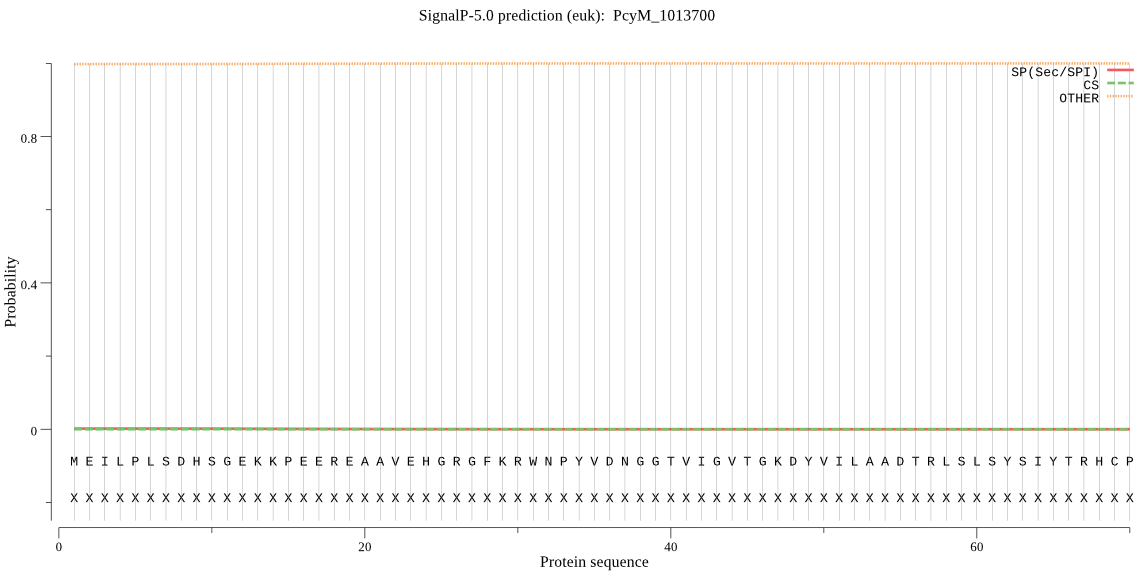
| PcyM_1013700 | OTHER | 0.999938 | 0.000052 | 0.000010 |
MEILPLSDHSGEKKPEEREAAVEHGRGFKRWNPYVDNGGTVIGVTGKDYVILAADTRLSL SYSIYTRHCPKISKLTDKCIIGSSGMQSDIKTLHSLLKKKIELFVLEHAHYPDIHVIARL LCVILYSRRFFPYYTFNLLAGVSNENKGVLYNYDAIGSYCEATHSCVGSGSALILPILDN RMEQKNQLIKNLNFNLRDDIDFVKDAITSATERDIYTGDKTEIYIIDHMGVNTTTLELKQ D
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_1013700 | 10 S | LSDHSGEKK | 0.991 | unsp |