

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
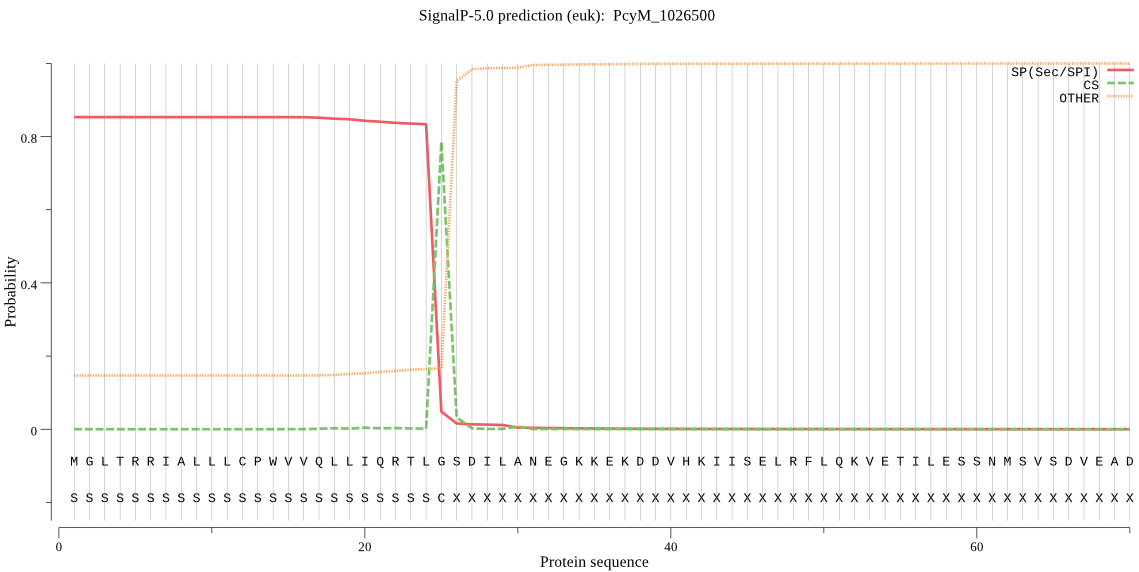
| PcyM_1026500 | SP | 0.177036 | 0.816652 | 0.006313 | CS pos: 25-26. TLG-SD. Pr: 0.9635 |
MGLTRRIALLLCPWVVQLLIQRTLGSDILANEGKKEKDDVHKIISELRFLQKVETILESS NMSVSDVEADVNAYNPDRDAPREELEKLQDQQEEPEKEINHLRKNPQRRTEKKEASIKNK KALRLIVSENHATSPSFFEESLLQEDVMSFIQSKGKLANLKNLKSIIIDLNDDMTDEELA EYISMLERKGALIESDKLVGADDISLASIKDAVRRGESRLNLDKFHSTMLEVQDGEAKTS HPGSAIGGSDVSDVNGGSDVRGGDSSSDDDILLEGSIHTESYLPGRKTSNSYKFNDEYRN LQWGLDLARLDETQDLINTNRVSVTKICVIDSGIDYNHPDLKGNIDVNMKELYGRRGVDD DNNGVVDDVYGANFVNNSGDPMDDNYHGTHVSGIISAVGNNNIGIVGVDGHSKLIICKAL DQHKLGRLGDMFKCIDYCISRKAHMINGSFSFDEYSSIFNASVNHLRSLGILFFVSASNC AHDKLTEPDITKCDLAVNYRYPPILSKTHNNVIAVANLKRDLDDSYSLSVNSFYSNIYCQ LAAPGTNIYSTTPFNSYRKLNGTSMASPHVAAIASIIRSINPNLSYVQIVEIMKNAIVKL PSLTEKVSWGGYVDILLAVNLAIDSKSVPYIKSQSWFRWKQRAR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1026500 | 116 S | KKEASIKNK | 0.993 | unsp | PcyM_1026500 | 116 S | KKEASIKNK | 0.993 | unsp | PcyM_1026500 | 116 S | KKEASIKNK | 0.993 | unsp | PcyM_1026500 | 134 S | NHATSPSFF | 0.992 | unsp | PcyM_1026500 | 208 S | ISLASIKDA | 0.997 | unsp | PcyM_1026500 | 265 S | RGGDSSSDD | 0.995 | unsp | PcyM_1026500 | 266 S | GGDSSSDDD | 0.992 | unsp | PcyM_1026500 | 291 S | KTSNSYKFN | 0.997 | unsp | PcyM_1026500 | 63 S | SSNMSVSDV | 0.996 | unsp | PcyM_1026500 | 65 S | NMSVSDVEA | 0.991 | unsp |