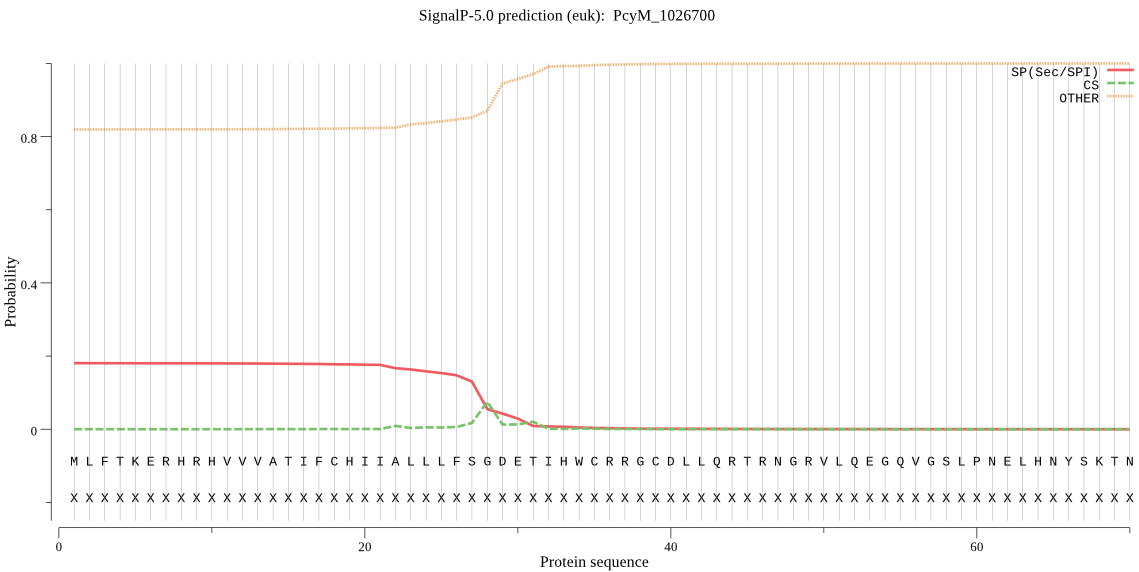
Fasta :-
MLFTKERHRHVVVATIFCHIIALLLFSGDETIHWCRRGCDLLQRTRNGRVLQEGQVGSLP
NELHNYSKTNLKIPSFAQMKLKDSHDRKIRRGGVTPEGNYHIPNCEDTDASSNFLKKFYR
SIKALVGITSHDTSWCLKYEKFRRFKQHFAYMVQSNLSVGKTRVCLIDTGVDLQDKVVGH
FVKIHKGEHTEGGGDNPSERSGGGDNPSERSGGGDNPSERSGGGEYGVHPQLEEQNYDGG
SPSWAYPKGRGTQYDGINTERCNEKNYDSCQSSDVDDVDMHGTFIANTVIRRDLLIKRGT
YKRGVELIVCKAFGDTEKTNSNLMPLIKCLEYCKERDAKVIHVGYNVQEESEKLVEVMKD
LEQAQIVVVSPSLQVYAAERGEAHAKKENGEEPSTHKMYPSSFADTFENVFSVGALRNSP
QGGLVPILDNANLPTGEKQKREQLHKRENTTLFSLSYGKVFPFGRSPSSMVEDGQGYASA
DFVNTLVMIFNVNPELSVKRMRLILERSIVRRSELKGLSKWGGYIDLFKVIGATLKERNQ
LCKTFFRELSGGGLPTGELPTGELSGGELSGGELSGGELSGGELSGGELSGGGLSGGELS
GGELSGGELSGGELSGGELSTGELSTGELSTGELSGGGMSSGGLSGGELSGGELSGGELS
GGELSGGELSTDGLSTDGLTGHLGDWAERPPPTGEEQPNSGEAITGEVTPRLEAASEEGE
EVTSRLEDQIVFPQHSGEMEEIERQLGGETLRGLGPHEEGNDMAQHDMAQYDLGYYDVEH
NEVEHNEVEHNEVEHNEVEHNDVEHYDLGYNDVEHYDLGYYDPDYYDTDVISKGAEDFSD
GVAALSEDPPSDTTYSGSYNAEEDVPDVPDMPDMPDMPDMPDMPDMPDMPDMPDEAKNSF
YGDTGGVTVAGGVSKLPLGVSFLDNHTSNEGNVPPLHRRNERGQVYGSGEGTSPPLNRRS
LGEKSGFPERWDQQGTHAMNEEDSDDGHHGMDEWVTQVKGNKYMDGSAYDVEKEHGNSRE
GIPAGELLRADWGWSRPPLSRWDQSGETSPWGDDKKGYDFSPQPDSNWGEKYSSSRRDPQ
RRRTRDLLGRKRAGNELGRRRRRPPRGMPRQRRRTPSRGRTTKRLKRREAGVVRKNGRTG
KSRSDHAMRSRSSRGYGNRTVRGRPVRGRPVRGNPVRGNPVRGNPVRAFAPKMSRVAMGR
R