

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
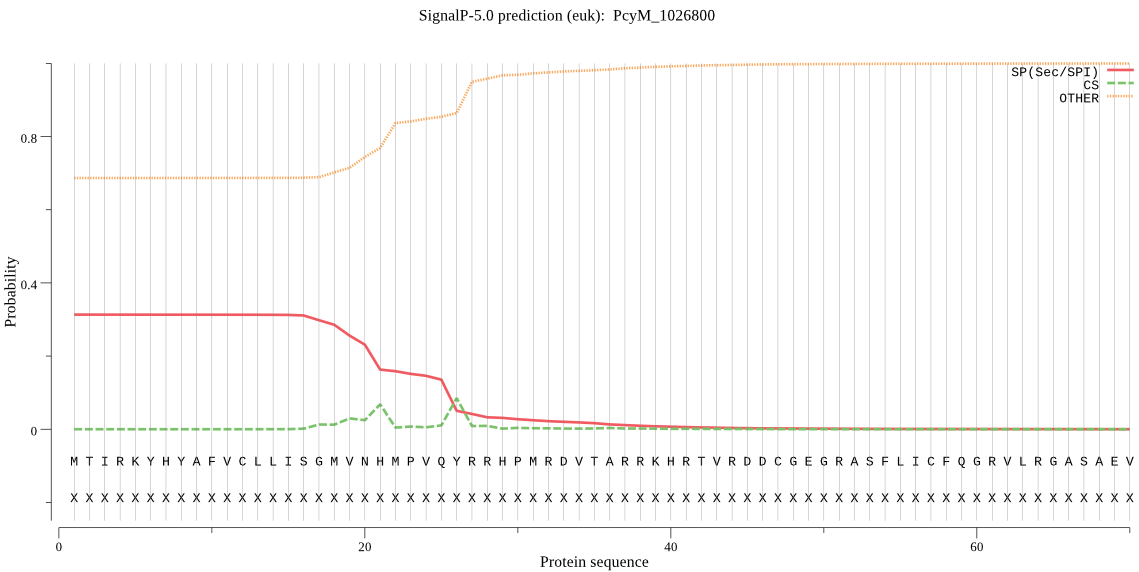
| PcyM_1026800 | OTHER | 0.765346 | 0.227587 | 0.007068 |
MTIRKYHYAFVCLLISGMVNHMPVQYRRHPMRDVTARRKHRTVRDDCGEGRASFLICFQG RVLRGASAEVLKVGGFQTGVGSSSLSSSPASSPSSPPPSSPPPSSPPSSPPSSPPSPSAH SPPRGDHLLQEDPHHMDETLTKLRKKKMIIKFKQQENASQSALIQQNIVNLLGSCGRVKK LSHIDLYLYETFSNISERALKNCLQLLSSGPVLVEQDFQIHAVERGGCMSGMVDKGGIVD VGDMADKGGIVDVGGLYAIRAIGGEAPARSGTNLRSHAGSNGTFQLNNQMAFKNFLSRLQ SNYKGTNIINGYDQTKMKEGTEMSEPHEQNDVNVCIVDTGVDYNHRDLRGNVVHVLHGRD VRGDGSGRGGSGRGGSGRGGSGKDGSGRDGSGTDGRSWDDGDRGDRPGGMDNHGHGTFIA GIIAGNSQRESQGINGICKRAKLTICKALNSRNAGVVSDILKCFNFCASKEAKIINASFA STKNYVSLFEALKTLEEKNILVVSSSGNCCPTTESMNTFPECNLDVKKVYPTAYSLNLRN LITVSNMIQQENGQVILSPDSCYSANYVHLAAPGDDIISTFPRNKYAISSGSSFSAAVVT GLAALVLSINARLSYEEVIDLLRGSIVQTESLRSKVKWGGFLDVRHLVSSTIALSRGEAE AARE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1026800 | 109 S | SPPSSPPSS | 0.993 | unsp | PcyM_1026800 | 109 S | SPPSSPPSS | 0.993 | unsp | PcyM_1026800 | 109 S | SPPSSPPSS | 0.993 | unsp | PcyM_1026800 | 116 S | SSPPSPSAH | 0.995 | unsp | PcyM_1026800 | 121 S | PSAHSPPRG | 0.997 | unsp | PcyM_1026800 | 182 S | VKKLSHIDL | 0.997 | unsp | PcyM_1026800 | 381 S | GRGGSGKDG | 0.998 | unsp | PcyM_1026800 | 386 S | GKDGSGRDG | 0.997 | unsp | PcyM_1026800 | 391 S | GRDGSGTDG | 0.997 | unsp | PcyM_1026800 | 397 S | TDGRSWDDG | 0.997 | unsp | PcyM_1026800 | 427 S | IAGNSQRES | 0.99 | unsp | PcyM_1026800 | 614 S | NARLSYEEV | 0.998 | unsp | PcyM_1026800 | 655 S | TIALSRGEA | 0.994 | unsp | PcyM_1026800 | 92 S | SPASSPSSP | 0.996 | unsp | PcyM_1026800 | 95 S | SSPSSPPPS | 0.99 | unsp |