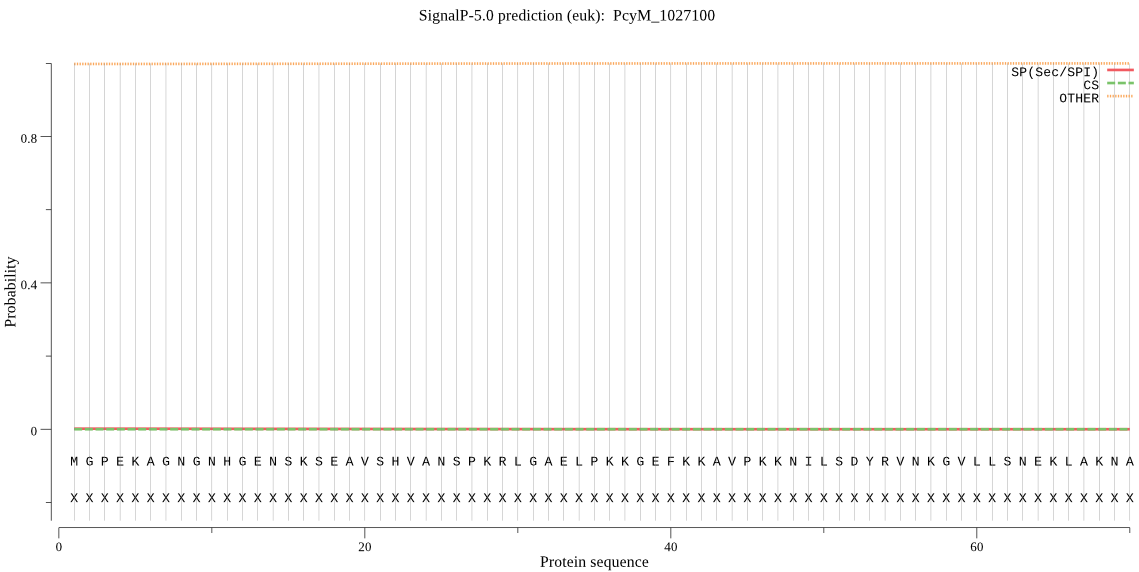
Fasta :-
MGPEKAGNGNHGENSKSEAVSHVANSPKRLGAELPKKGEFKKAVPKKNILSDYRVNKGVL
LSNEKLAKNADSSDREVASGGNKEDSDEESDLNITVGRGGGTRSKRDTTAAAGETATTAA
ITTTATAAKKGRNGPSDIKILVTNDEPLHTLPSGAVGRRAPLNPFSSPILGKYRRKNKNA
KIKVKDPRLNNNPLVGRLVVCISSTAILFWIFFAEMIFNYNTFNGRCISKVLYPIYTESV
MKKREPFFVFLGYGACEYNLEESASDRHFVGTSASDKGWPTNKVEENRDARGESSWDSVN
TRVYNQLGGLNTNYIRNYGEIYRLFWSVYLHGGFMHIIFNVICQIQILWIIEPDWGFIRT
LILFFTSGVTGNLLSAVCDPCGVTIGSSGALYGLIGALFTYYIEYWKTIPRPCCVVIFML
IVTIFGIFIGMFGYTDNYAHMGGCLGGILYGFATITTVSAADKCTLGERMLTSAPFSWFL
SDETKELINAKAREKKIRGDNYRKKQIANKVHKNDALHVMMSLMKNRINEEGRPPCKMKL
REWIVRITAASALIILWIVLFVYLLNETAYKSYTPMGQIKFTGVHTCFCCEVNKDKFPFI
SAEKFYWCFTNEEATKYYCGK