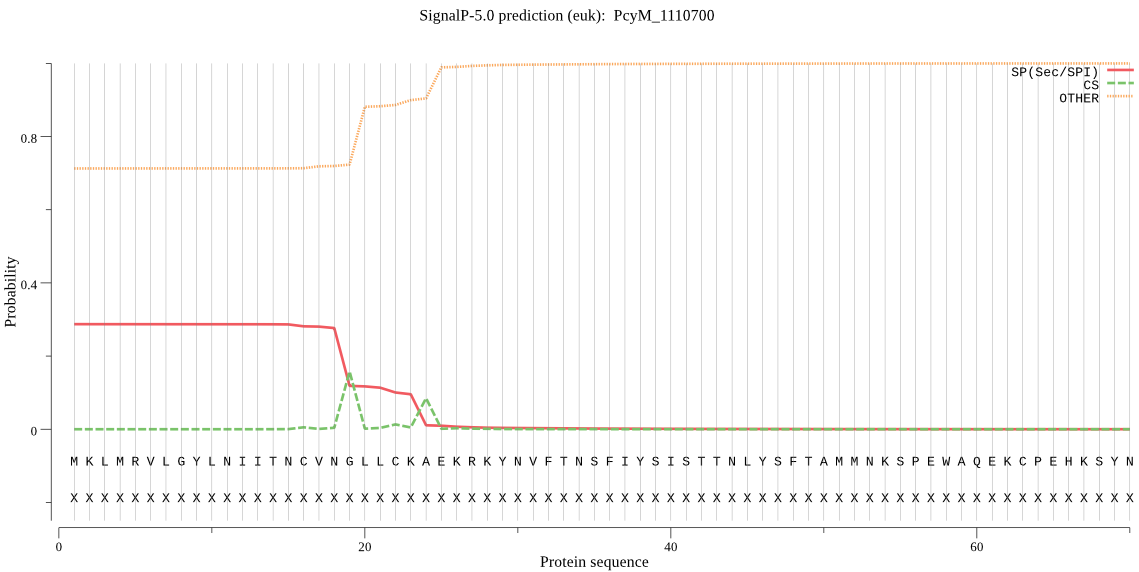
Fasta :-
MKLMRVLGYLNIITNCVNGLLCKAEKRKYNVFTNSFIYSISTTNLYSFTAMMNKSPEWAQ
EKCPEHKSYNILEKRFSEKFKMTYTVYEHKKAKTQVIALGSNDPLDVEQTFAFYVKTLTH
SGKGIPHILEHTVLSGSKNFNYKDSMGLLEKGTLNTHLNAYTFNDRTVYMAGSMNNRDFF
NIMAVYMDSVFQPNVLENKFIFQTEGWTYEVEKLKDEEKNTDAPKIKDYKVSYNGIVYSE
MKGSFSSPLQHLYYVIMKNTFPDNVHSNISGGDPKEIPTLTYEEFKEFYYKNYNPKKIKV
IFFSKNNPTELLNFVDDYLNQLDFTKYRDDAVENVNYQEYKKGPFYVRNKFADHSEEKEN
LVSISWLLNPKKNDLLDVDLSLESPQDYFALLIINNLLTHTTESVLYKALIDCGLGNSVI
DTGLDDSLVQFIFTIGLKGIKEKNEKNISLDKIHYEVENVVLKALQKVVDEGFNKSAVEA
SINNIEFVLKEANLKTSKSIDYIFEMASRLNYNRDPLLIFEFEKHLNVVKDKIKNQPKYL
EKFIEKHFINNYHRTVILMEGDENYGKEQEELEKESLKKQIESLTEKERDDIIVDFENLT
KYKNTVESPEHLDNFPIISISDLNKETLEIPANAYFTTISEENNMDKYNQVKSSEDLMKK
NMDDLINKYVLKGSQGEAITDGATKQSNVAEGEIPMLIYEMPTSGILYLQLIFSLDHLTL
EELSYLNLFKVLILENKTNKRSSEEFVILREKNIGNIMANVALYSTSDHLKVTSKYNAHG
LFNFEMHVLSHKCNESLEIALEALKDSDFSNKKKIVEILKRKINGMKTVFSSKGYSLLLK
YVKSQLNAKYYAHDLVFGYGNYLKLQEQLKLAESDFDKFEEILNRIRNKIFTKKNLLISV
TSDSAALDELFVKSKESFKNLLGYFEQNDSKNNGTETIGWIEEIKQSKVIEKEQKKKEFF
VIPTFVNAVSMAGILFNEKEFLDPSFIVIVAALKNSYLWETVRGLNGAYGVFADIEYDGA
VVFLSARDPNLEKTLQTFRESAQGLRKMADTMTKNDLRRYIINAIGNIDKPRRGVELSKL
SLLRIISNETKQDRIDFRKRIMETTKEDFYKFADLLEKKINEFEKNIVIITSKEKASGYS
TNVDQDFKLIHIE