

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_1113300 | SP | 0.003859 | 0.974947 | 0.021195 | CS pos: 18-19. GNA-FL. Pr: 0.8813 |
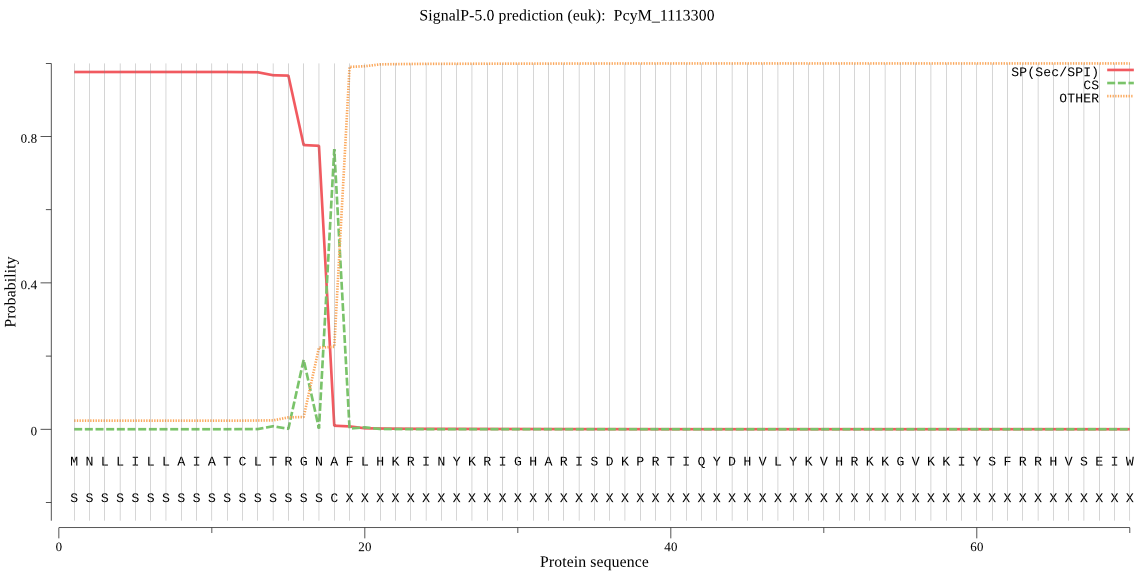
MNLLILLAIATCLTRGNAFLHKRINYKRIGHARISDKPRTIQYDHVLYKVHRKKGVKKIY SFRRHVSEIWNNIKTRRIENYMEDFFQTEQVKNTLQNNFLCFKHLFNKCKLDRVIIAVNV ALYFYLNRIDKDDEKKIFFHKGNLLEVKEQKAEKYKCNYHDVYKTKNFKTFFTSLFIHKN ILHLYFNMSSLISIYKLISLIYTNSQIFVTYLLSGVLSNIISYICYLREKKENVLLKNLI LKNASNENNVLINKNNKIICGSSSCIYSLYGMHMTYIIFFYFKHNYIVNSNFLYNFFYSF VSSLLLENVSHLNHMLGFLCGFFFSSLIVLFDRNA