

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
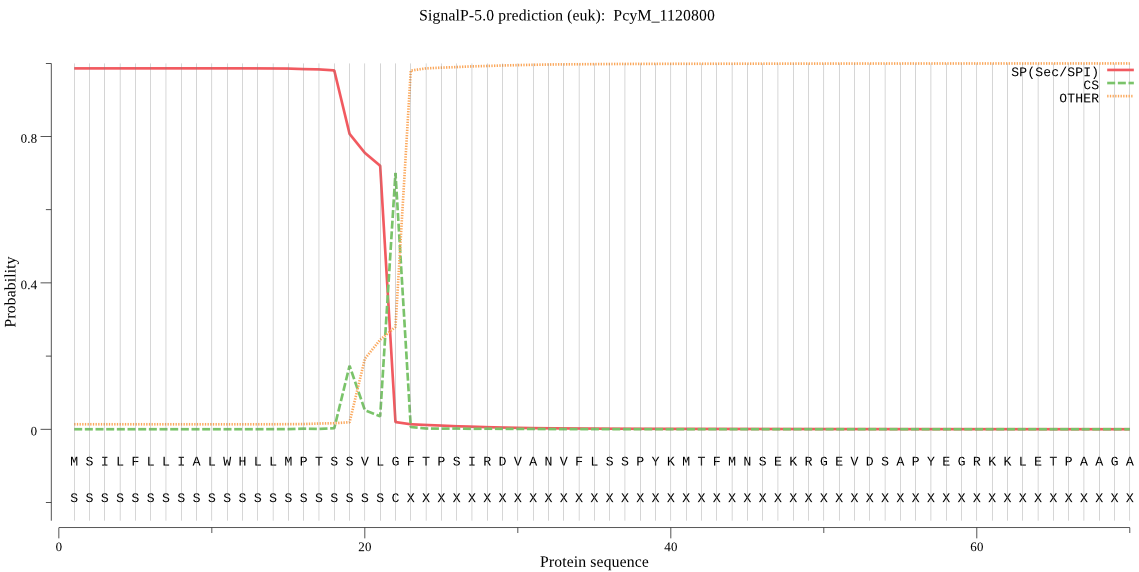
| PcyM_1120800 | SP | 0.007974 | 0.991922 | 0.000104 | CS pos: 22-23. VLG-FT. Pr: 0.6370 |
MSILFLLIALWHLLMPTSSVLGFTPSIRDVANVFLSSPYKMTFMNSEKRGEVDSAPYEGR KKLETPAAGAEREEATVRVVQAEPEGEKTQGGEEEKKAEAPKTGVATDGKAEEVAATEDV AATTQEVAATEEAATTQEVAATEEAATTQEVAATEEAATTQEVAATEEAATTQEVAATED VATTQEVAATEDVATTQEVAATEDVATTQEVAATKDVATTQEVAATKDVAATEEAATTQE VAATKEVAATKEVAATKEVATTKEVAATKEVAATKEVAETKEVAATKEVAATKEVAATEE AAVTEEAAVTEEAATTQEVAATEEAVTVEELSSTVEEAETTEEAATVEELETTEEAETTE GEAAPAEVSERMRKKAKGKSNERAAHVGNILKKKKYKLFSKQKPNKNKKKEKKKKEPKDK SKKKDREKLPTTFLLPGVGGSTLIAEYKNALIHSCSNNLLNSKPFRIWISLTRLFSITSN VYCTFDTLRLLYDNEKKMYFNQPGVNITVENYGRLKGIDYLDYINNTGIGVTKYYNTITS HFLSKGYVDGESIMGAPYDWRYPLHQQDYNLFKDSIEAAYERRNGMKVNVVGHSLGGLFI NYFLVHIVDKEWKQKYLSSIMYMSSPFKGTVKTIRALLHGNRDFVSFKIKNLIKLSISDS MMKAIGNSVGSLFDLIPYKEYYDHDQVVIIMNTDPTPIDKNNMQSIVTTCGIYNKDCYLN RTDVKLKVYTLSDWHVLLKDDLREKYNNHKQYRERHFSMDHGVPIYCVYSTLKNKSTDYL LFFQKENLNEEPDIYYGIGDGTVPIESLEACNNFYNATEKKHFENYSHIGILHNADTVKY VYDSLQTIGGNLDDVREDVTAKVALQGSVEGEGKEQMLPQGSNGAENELSKVPQQGGGNG EMTNAEGQRAK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1120800 | 421 S | PKDKSKKKD | 0.996 | unsp | PcyM_1120800 | 421 S | PKDKSKKKD | 0.996 | unsp | PcyM_1120800 | 421 S | PKDKSKKKD | 0.996 | unsp | PcyM_1120800 | 46 S | TFMNSEKRG | 0.994 | unsp | PcyM_1120800 | 380 S | AKGKSNERA | 0.992 | unsp |