

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_1226200 | OTHER | 0.986782 | 0.013132 | 0.000086 |
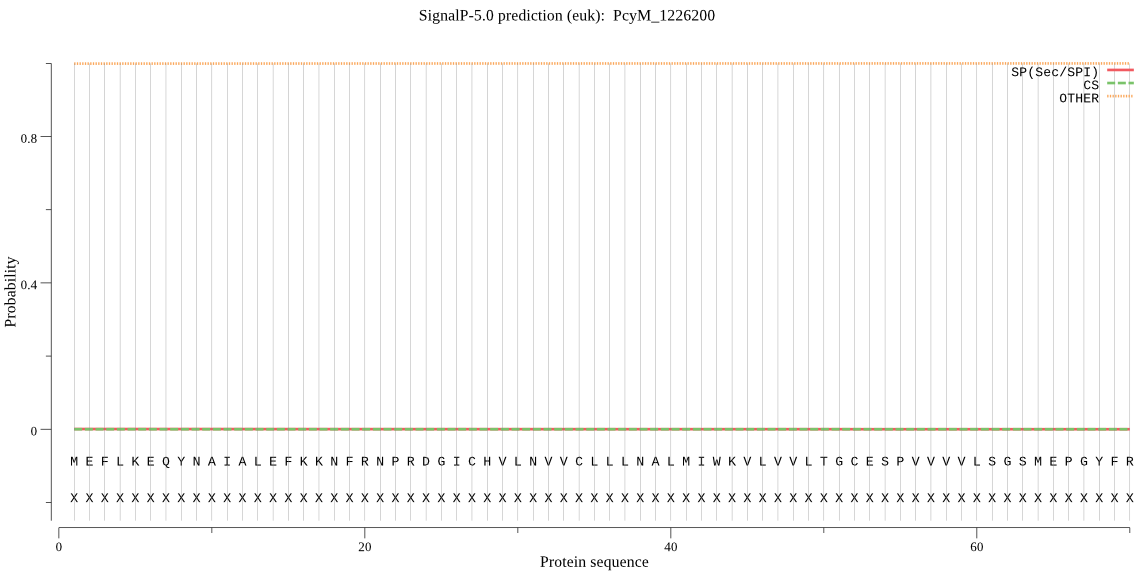
MEFLKEQYNAIALEFKKNFRNPRDGICHVLNVVCLLLNALMIWKVLVVLTGCESPVVVVL SGSMEPGYFRGDTLALWHPPNIHAGDVVVYQINGRDIPIVHRILNIHISKDNKFHLLSKG DNNNIDDRGLYEPNQYWLENEHVLGLSVGYAPYIGMLTIWVNEYPVMKWGIVSLMLFMIL LGYE