

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
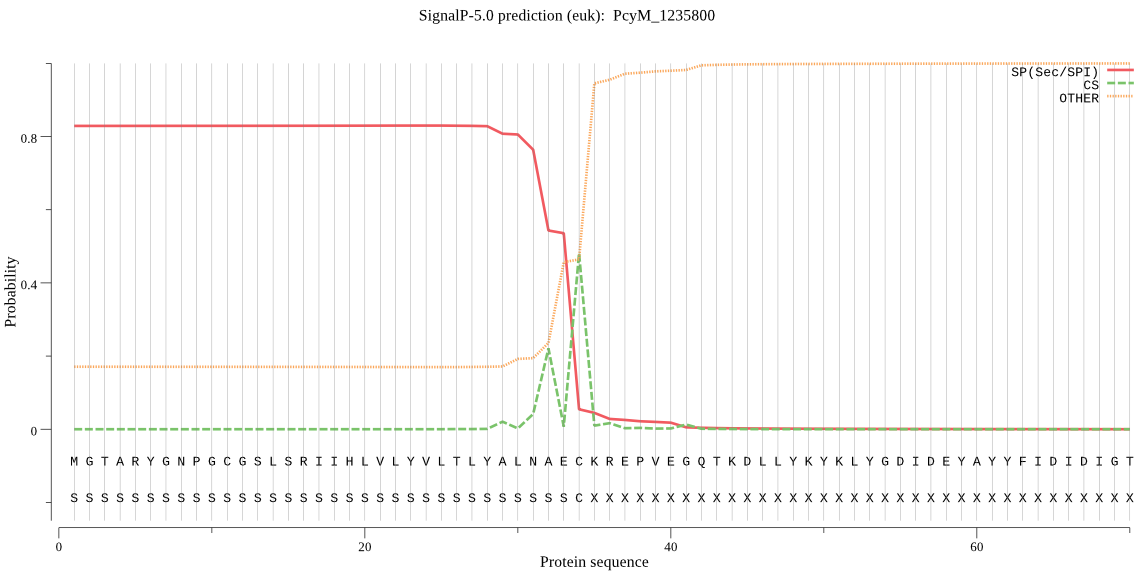
| PcyM_1235800 | SP | 0.008091 | 0.991900 | 0.000009 | CS pos: 34-35. AEC-KR. Pr: 0.7751 |
MGTARYGNPGCGSLSRIIHLVLYVLTLYALNAECKREPVEGQTKDLLYKYKLYGDIDEYA YYFIDIDIGTPEQRISLILDTGSSSLSFPCAGCKNCGVHMENPFNLNNSKTSSILYCDEN EECPFKLNCVKGKCEYMQSYCEGSQISGFYFSDVVSVMSYNNEKISFRKLMGCHMHEESL FLYQQATGVLGMSLSKPQGIPTFINLLFGNAPQLKEVFTICISENGGELIAGGYDPAYIV RKGKEQSQGQGQSLGQGQSQGKEQAAQQGEPKQRGGDSPTREDPELALRDAEKIVWESVT RKYYYYIRVRGLDLFGTNMMSTSKGLEMLVDSGSTFTHIPEDLYNKLNYFFDILCIQDMN NAYDVNKRLKITKESLNNPLVQFDDFRKSLKTIISKENMCVKIVEGVQCWKSLEGLPDLF VTLSKNYKMKWQPHSYLYKKESFWCKGIEKQVNNKPILGLTFFKNRQVIFDLQKNRIGFV DSNCPSHPTHTRPRTYNEYKRKDNIFLKIPFFYLYSLFVVFALSILLSLVFYVKRLYHME YNALPSEGKAPADA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1235800 | 278 S | RGGDSPTRE | 0.993 | unsp | PcyM_1235800 | 389 S | DFRKSLKTI | 0.994 | unsp |