

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PcyM_1262900 | SP | 0.028135 | 0.971810 | 0.000055 | CS pos: 21-22. SKA-NV. Pr: 0.7658 |
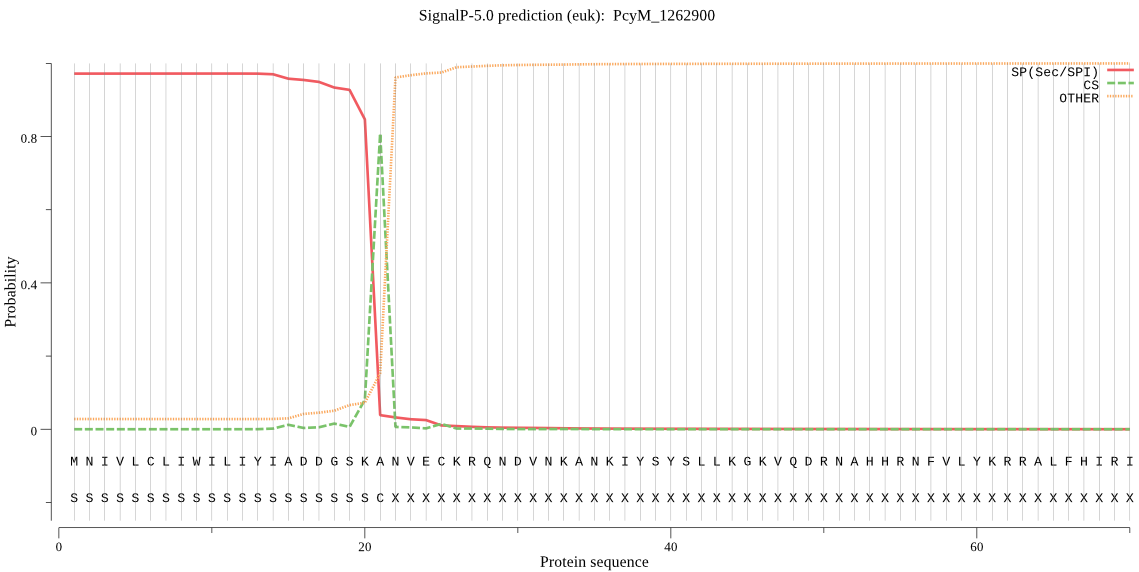
MNIVLCLIWILIYIADDGSKANVECKRQNDVNKANKIYSYSLLKGKVQDRNAHHRNFVLY KRRALFHIRIPRKNLERHNLFSNDLRYKKINKFLENKKTNKLCYLHISNNHHGIKKKRNK NKTLQLLFEKKKLLQEYFCNLKSSFMDRYRNTKIVTKLFLSSSLLMLTLNLIGVKPEDIA LHDKRIIRAFEFYRIVTSALFYGDISLYVLTNVYMLYVQSQELERSVGSSETLAFYLSQI SILSVICSYLKKPFYSTALLKSLLFVNCMLNPYQKSNLIFGINIYNIYLPYFSIFIDILH AQDLKASLSGILGVTSGSIYYLLNIYAYEKFNKKFFLIPRFLRNYLDSVSTDDVF