

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
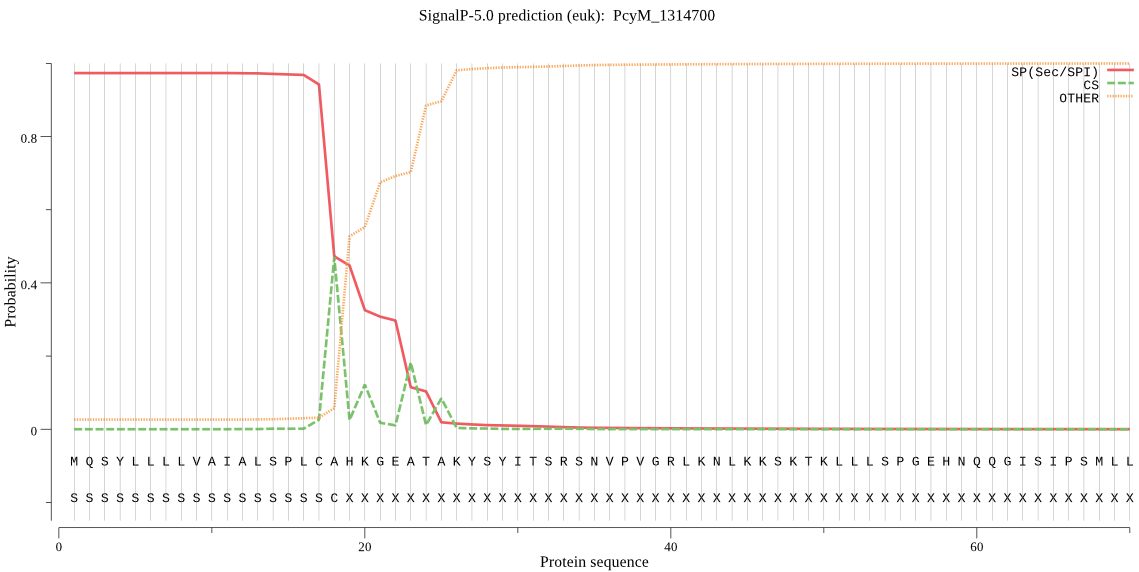
| PcyM_1314700 | SP | 0.001418 | 0.998372 | 0.000209 | CS pos: 23-24. GEA-TA. Pr: 0.5855 |
MQSYLLLLVAIALSPLCAHKGEATAKYSYITSRSNVPVGRLKNLKKSKTKLLLSPGEHNQ QGISIPSMLLSKRIIFLSSPVYPHISEQIISQLLYLEYESKRKPIHLYINSTGDLENNKI INLNGITDVISIIDVIDYISSDVYTYCLGKAYGISCILASSGKKGFRFSLKNSSFCLNQS YSVIPFNQASNIEIQNKEIMNTKRKVVEIIANNTGKEKSHIERILERDRYFSAPEAVQFN LVDHILEKE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1314700 | 232 S | DRYFSAPEA | 0.996 | unsp | PcyM_1314700 | 232 S | DRYFSAPEA | 0.996 | unsp | PcyM_1314700 | 232 S | DRYFSAPEA | 0.996 | unsp | PcyM_1314700 | 54 S | KLLLSPGEH | 0.994 | unsp | PcyM_1314700 | 169 S | GFRFSLKNS | 0.996 | unsp |