

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
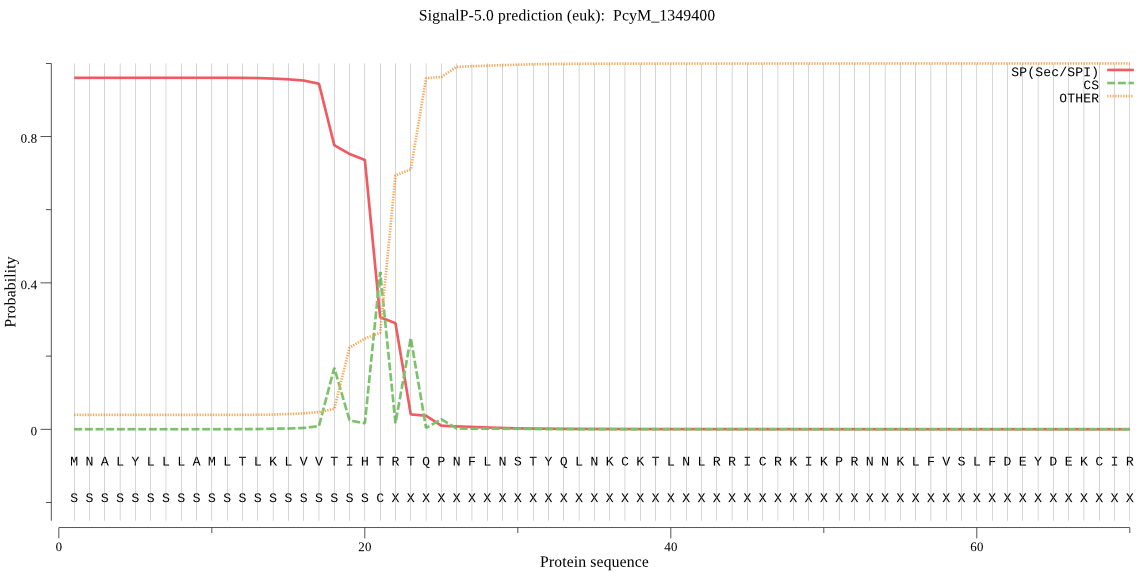
| PcyM_1349400 | SP | 0.071124 | 0.923220 | 0.005656 | CS pos: 21-22. IHT-RT. Pr: 0.6343 |
MNALYLLLAMLTLKLVVTIHTRTQPNFLNSTYQLNKCKTLNLRRICRKIKPRNNKLFVSL FDEYDEKCIRALIMAREVAKNQNEKEIQLKHLLIAIIRIDSNLVKHILNFFNISITTFLD KLNKEMNNRRGNTSFGTNQDKQNASPGEGTPIQEIPIQETPLQETPLQETPLQETPTDQL PPPDNSNSIVEGERSHQSNDPNGEHKLMNAFINKHIQDIEDKINQLKNLDKGQSDISTEV DDADSSVEGSNERDEAARNLIRDAAKRVLGDHAGDLEDPPGGPPLLDQATPGELPTDLPS DLPSDVSKELSKELSKELSKELSKELSRQLPSDLANNLPNDLPDEKADIKFSQNCKRVLH NAVLEARRKKKMFVNVTDILIALINMAQENQNCEFLRYLSELNISVNDLKRELTTYDEQN CFAPPTGRASRNSQRDDDQNTNHTEKREENGSHRETSENRDENQRMRSMNNEQTNHHFMN NLNNDYLNQSREFKNYEEGSFPPNNKLFSSSYVKDCLTDMVQAAYEKGDEHFFGRKKEIK RIIEILGRKKKSNPLLIGESGVGKTAIIEHLSYLILKDKVPYHLRNCRIYQLNVGNIVAG TKYRGEFEEKMKHLLNNMNKRKKNILFIDEIHVIVGAGSGEGSLDASNLLKPFLSSDNLQ CIGTTTFQEYTKYIESDKALRRRFNCVPVKPFTAKETLLLLKKIKYNYEKYHNIYYTNDA LKSIVMLTEDYLPTANFPDKAIDILDEAGAYQKIKYEMFMRQRLRDERAGQVGGNQGSGN QGSGGQGSGGQGSGDQVITRQVNAAHLSGDEHPPAKLHTADSEPADIDEDRNVEAQNYLQ NMHMKYVTSDVIENIVSKKSSISYIKKNKKEEEKIIKLKEKLSKIIIGQEKVIDILSRYL FKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVISKYLFNEDNLIVINMSEYIDKHSVSK LFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHSEVLHVLLQILDNGILTDSKGN KVSFKNTFIFMTTNVGSDIITDYFKLYNKNYSNLGFKYYLSKSQNREDTHSSSLSKGQLP EGVNSVGNSVVEHPTRLSNNGGADIQIDHKTYDHPVENPPNQQPNSYEIFEEKLRTNKWY EELQPEIEEELKKKFLPEFLNRIDEKIIFRQFLKRDIVNIFQNMIDDLKKRIKKRKNLNL IIEQGVIKYICSDENNIYDMNFGARSIRRALYKYIEDPIASFLISNLCEPNDSIHVSLSS DNKINVQLLKASVDQLVL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1349400 | 250 S | SVEGSNERD | 0.99 | unsp | PcyM_1349400 | 250 S | SVEGSNERD | 0.99 | unsp | PcyM_1349400 | 250 S | SVEGSNERD | 0.99 | unsp | PcyM_1349400 | 430 S | TGRASRNSQ | 0.99 | unsp | PcyM_1349400 | 433 S | ASRNSQRDD | 0.998 | unsp | PcyM_1349400 | 452 S | EENGSHRET | 0.997 | unsp | PcyM_1349400 | 457 S | HRETSENRD | 0.994 | unsp | PcyM_1349400 | 145 S | KQNASPGEG | 0.996 | unsp | PcyM_1349400 | 237 S | QSDISTEVD | 0.994 | unsp |