

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
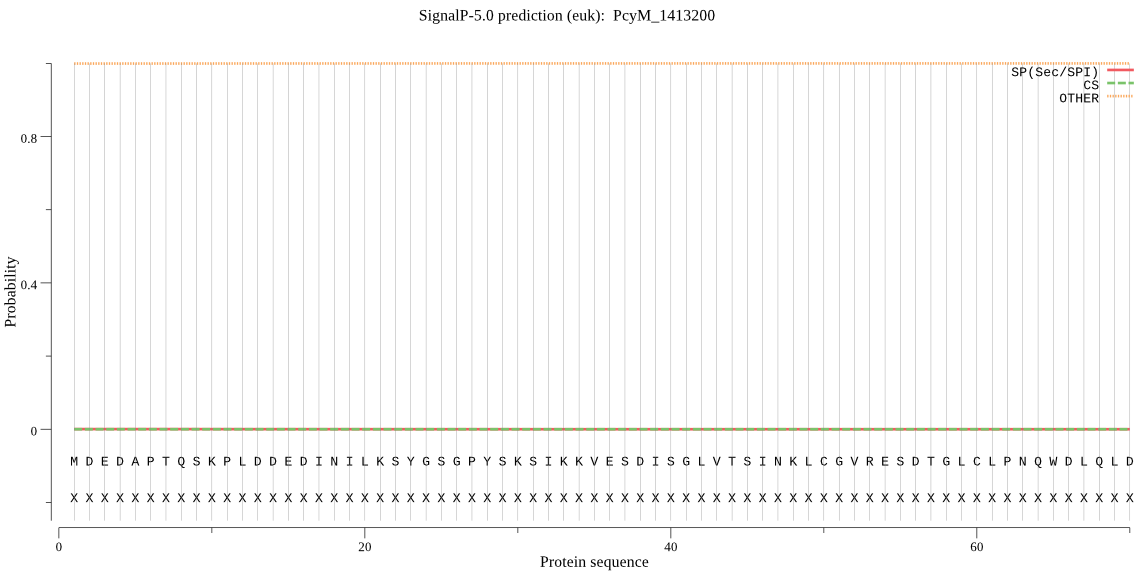
| PcyM_1413200 | OTHER | 0.999928 | 0.000033 | 0.000039 |
MDEDAPTQSKPLDDEDINILKSYGSGPYSKSIKKVESDISGLVTSINKLCGVRESDTGLC LPNQWDLQLDKQMLNEEQPLQVARCTKIINGDTDQTKYIINVKQIAKFVVGLGDKVAPSD IEEGMRVGVDRTKYKIQILLPPKIDPTVTMMTVEEKPDITYNDIGGCKEQLEKLREVVEM PLLQPERFVTLGIDPPKGVLLYGPPGTGKTLTARAIANRTDACFICVIGSELVQKYVGEG ARMVRELFQMAKSKKACILFIDEVDAIGGSRGDESAHGDHEVQRTMLEIVNQLDGFDNRG NIKVLMATNRPDTLDSALVRPGRIDRKIEFSLPDLEGRTHIFKIHANTMNMSRDVRFELL ARLCPNSTGSDIRSVCTEAGMFAIRARRKTITEKDLLLAINKVIHGCKQFSATGKYMVYN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_1413200 | 119 S | KVAPSDIEE | 0.993 | unsp |