

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
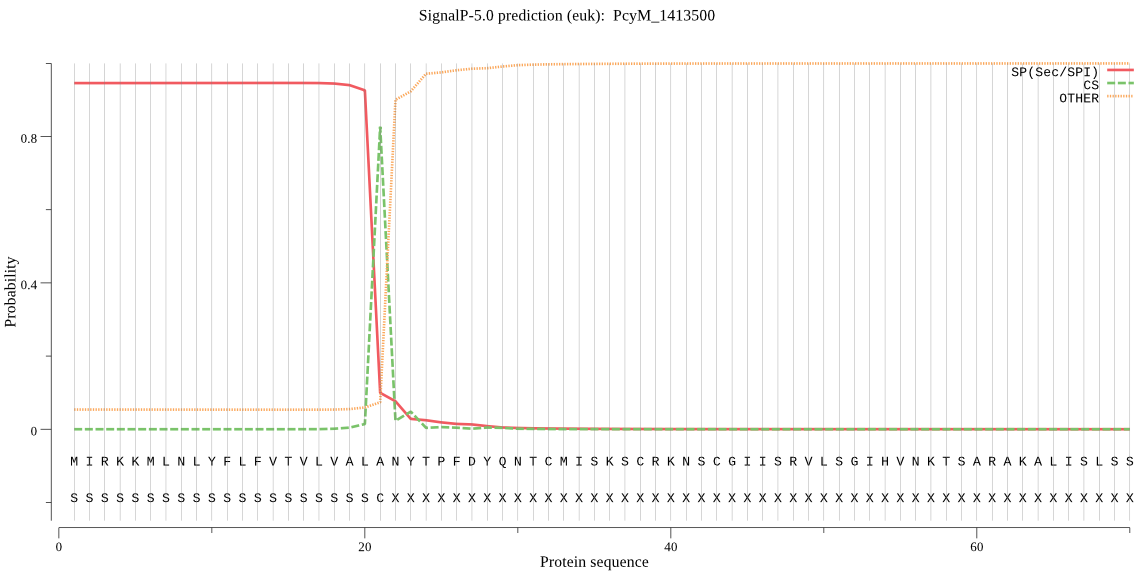
| PcyM_1413500 | SP | 0.053916 | 0.946028 | 0.000056 | CS pos: 21-22. ALA-NY. Pr: 0.8490 |
MIRKKMLNLYFLFVTVLVALANYTPFDYQNTCMISKSCRKNSCGIISRVLSGIHVNKTSA RAKALISLSSLIYQLQLPKLVSLDLFRRDLFTGVKQKGKRSPVPSYIIQNRLMSENIDSG STNMSVTANQDKKRPGTGDGSEGNNQGGSSTSATDKRFKEMDKNEESSNVSGSTNATLTN GTNSTTDGGENNNNGSGNQGKNEPTIHYRKDYKPSGFVIDNVTLNINIFDNETSVRSTLD MKLSEHYRGEDLIFDGVSLEIKEISIDGNKLKEGEHYKYDNEFLTIYSKFIPKGKFTFGS EVIIHPETNYALTGLYKSKNIIVSQCEATGFRRITFFIDRPDMMAKYDVTVTADKEKYPV LLSNGDKLNEFDIPGGRHGARFNDPHLKPCYLFAVVAGDLKHLSDNYVTKFSKKKVELYV FSEEKYVSKLKWALECLKKAMKFDEDYFGLEYDLSRLNLVAVSDFNVGAMENKGLNIFNA NSLLASKKKSIDFSFERILTVVGHEYFHNYTGNRVTLRDWFQLTLKEGLTVHRENLFSEQ TTKTATFRLDHVDILRSVQFLEDSSPLAHPIRPESYVSMENFYTTTVYDKGSEVMRMYQT ILGDEYYKKGMDIYIKKNDGGTATCEDFNDAMNEAYKLKKGDKTANLDQYLLWFSQSGTP HVTAEYSYDAGKKEFVIEVTQVTNPDPNQKEKKALFIPIRVGFINPHNGQDVIPEVTLEF KKDKEKFVFNNVNEKPIPSLFRGFSAPVYIKDNLTDAERILLLKYDTDAFVRYNVCVDLY MKQILMNYQELLKAKTENKQESDEKPSLTPVSEDFINAIKYLMEDPHADAGFKSYIITLP RDRFIINYIKNVDTDVLADTKDFIYKQLGDKLNDLYFQIFKSIQAKADDMTHFEDESYVD FEQLNMRKLRNTLLTLLSRAKYPNMLDQIMEHSKSPYPSNWLASLAVSAYYDKYFDLYEM TYNQSKDDELLLQEWLKTVSRSDRKDIYDIIKKLETEVLKDSKNPNEIRAVYLPFTYNLR YFNDISGKGYKMMADIIMKVDKFNPMLATQLCDPFKLWNKLDQKRQDMMLNEMNRMLSME NISNNLKEYLLRLTNKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1413500 | 237 S | TSVRSTLDM | 0.99 | unsp | PcyM_1413500 | 237 S | TSVRSTLDM | 0.99 | unsp | PcyM_1413500 | 237 S | TSVRSTLDM | 0.99 | unsp | PcyM_1413500 | 426 Y | SEEKYVSKL | 0.993 | unsp | PcyM_1413500 | 802 S | NKQESDEKP | 0.994 | unsp | PcyM_1413500 | 898 Y | EDESYVDFE | 0.992 | unsp | PcyM_1413500 | 1078 S | NRMLSMENI | 0.995 | unsp | PcyM_1413500 | 152 S | GSSTSATDK | 0.993 | unsp | PcyM_1413500 | 184 S | NGTNSTTDG | 0.996 | unsp |