

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
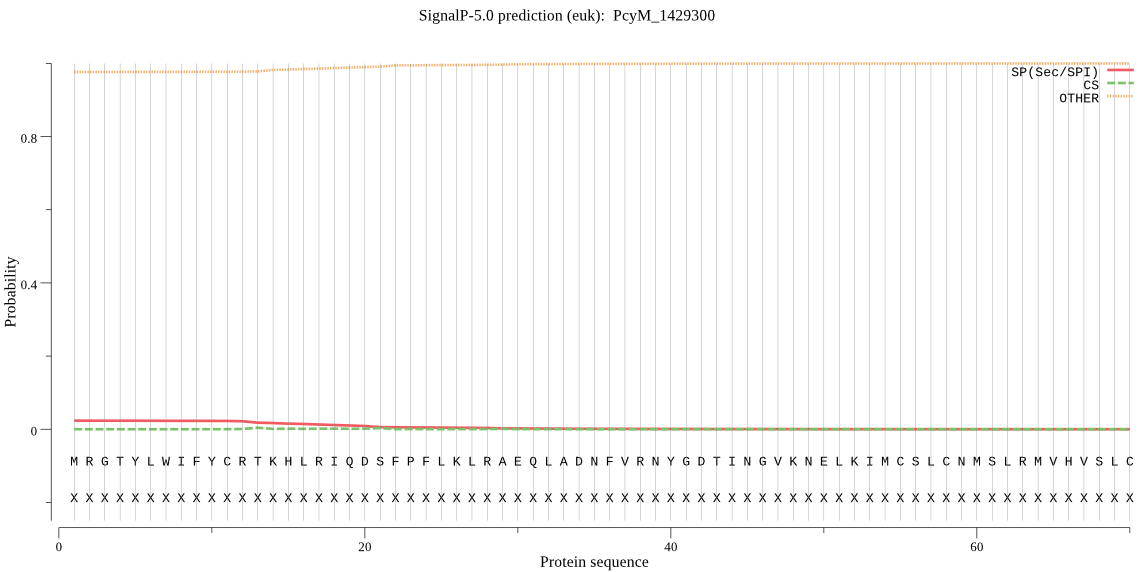
| PcyM_1429300 | OTHER | 0.987634 | 0.011755 | 0.000612 |
MRGTYLWIFYCRTKHLRIQDSFPFLKLRAEQLADNFVRNYGDTINGVKNELKIMCSLCNM SLRMVHVSLCESENSGGERTADGCERSFDCSFDRTFDRSSERTFDRSSECTFDRSSECTF DRSSECTFDRSSECTFEHSPEHRLSDGDHDAPAESDQLEQNTQPEKNDRSKLATHLIIHY VKEYVKYVAHSLLHFYFYLCDTFYRTLSVLSKSYLEFPKQFDYYASISDNLMTSFVTIYK IYKQSNSSEFCFKLDQLAFLGSGFIYDKRGYVLTAAHNITNTEGTFVIKNEDNFYVATLA GLHRESDICVMKINSEEQFSHIPLNSSREFLKPGEPVITYGQIQNFDKETYSIGVVNQPR QTFTKFENFCEKEQTCLYPFIQISNPINKGMSGSPLIDKHGNLVGMIQKKIDNYGLALPV NILKNITTHLQEEGTYKEPFLGIVLREKEQSTSAVYKNCKNELKIEDVLVNSPADVAGIT KGDIMLTINNTGIKNICDVHEILNSTSDGYVTVDIIRHGKNKKIKVKM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1429300 | 107 S | TFDRSSECT | 0.995 | unsp | PcyM_1429300 | 107 S | TFDRSSECT | 0.995 | unsp | PcyM_1429300 | 107 S | TFDRSSECT | 0.995 | unsp | PcyM_1429300 | 108 S | FDRSSECTF | 0.99 | unsp | PcyM_1429300 | 116 S | FDRSSECTF | 0.99 | unsp | PcyM_1429300 | 124 S | FDRSSECTF | 0.99 | unsp | PcyM_1429300 | 145 S | EHRLSDGDH | 0.998 | unsp | PcyM_1429300 | 99 S | TFDRSSERT | 0.997 | unsp | PcyM_1429300 | 100 S | FDRSSERTF | 0.996 | unsp |