

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
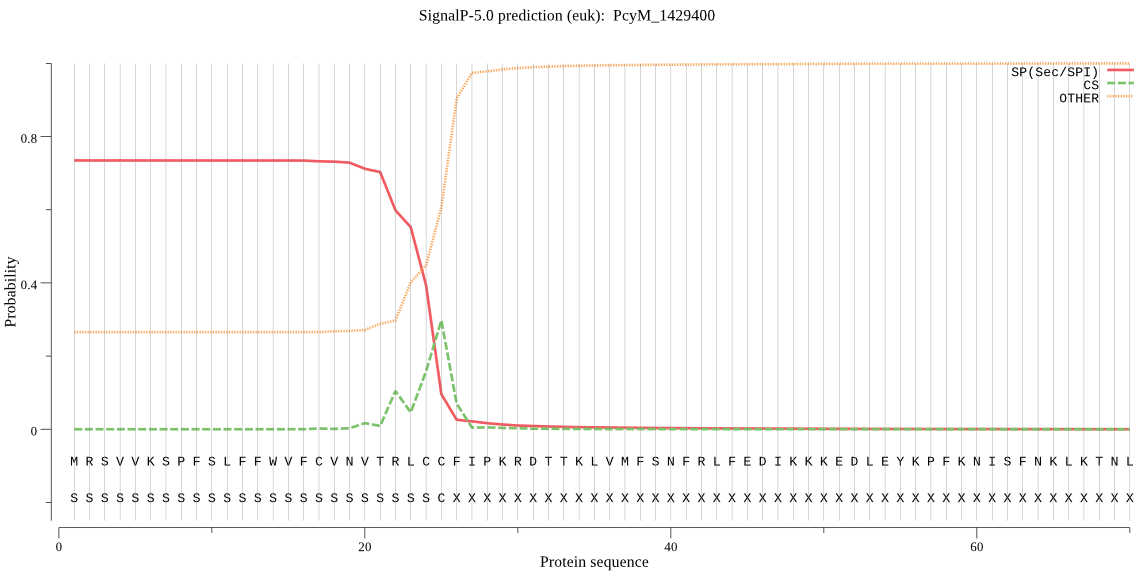
| PcyM_1429400 | SP | 0.117335 | 0.875108 | 0.007557 | CS pos: 25-26. LCC-FI. Pr: 0.4827 |
MRSVVKSPFSLFFWVFCVNVTRLCCFIPKRDTTKLVMFSNFRLFEDIKKKEDLEYKPFKN ISFNKLKTNLKNFLVINKKFNISDVKQVSFLGKFYNYEKIENYGVNEICILGRSNVGKST FLRNFIKYLINVNEHATVKVSKNSGCTRSINLYSFENGKKKRLFILTDMPGFGYAAGIGK KKMEFLRKNLEDYIFLRNQICLFFVLIDMSVDIQKIDVSIVDAIKKTNIPFRVICTKSDK FSTNAEERLQAIKNFYQLEKIPIHISKFSTHNYINIFKEIQHHCNLDTP
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_1429400 | 32 T | PKRDTTKLV | 0.99 | unsp |