

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
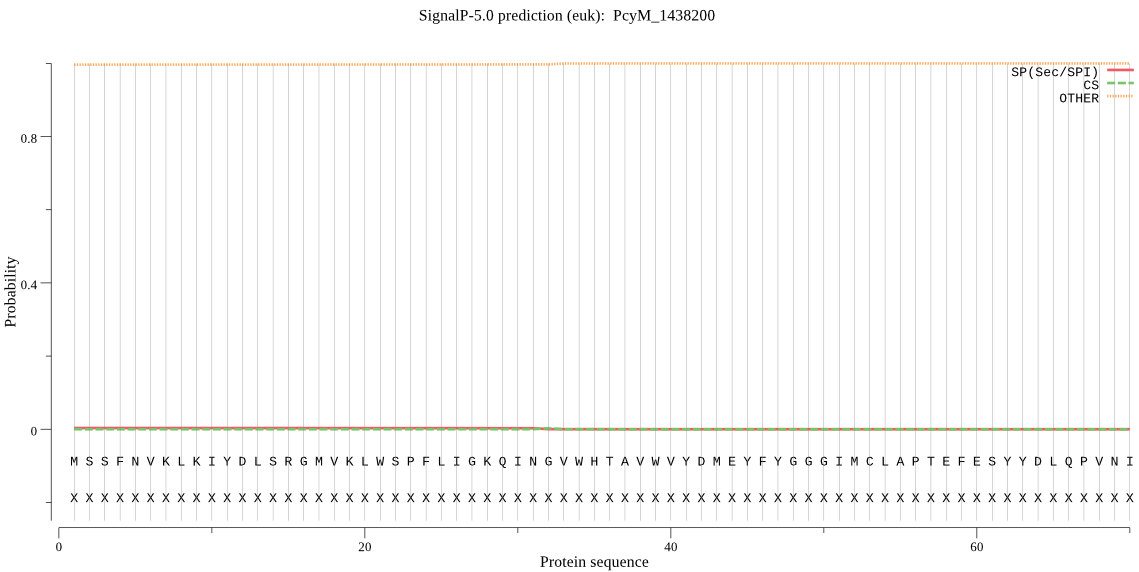
| PcyM_1438200 | OTHER | 0.993028 | 0.005317 | 0.001655 |
MSSFNVKLKIYDLSRGMVKLWSPFLIGKQINGVWHTAVWVYDMEYFYGGGIMCLAPTEFE SYYDLQPVNIVDMGITELQQSHFHEYLNGIQPDFTVDKYNLVNWNCNNFTNEVCNFLVGK NIPQYILDTPKEVMSTTKGKLILDMMQSYQTNIAPGFEASAPVLNNGRNEDSRGANIRAG CSKEAEVKELTKVPSVSLDNFFNENKIENLLDEYIKNEKYDVDEKKDFLNFLKNILDNVS TNPDVLKNRYVHVKNFPKFQSKHGNSEYNKILSSLNFFQGYIENDEVNNLKKFTIFVDPK YNKMHAFTEHLDLFLNKNIFLKNPDKTIYKMETLKFKDMCEYAAYCNKKKSNSNDVLIFI SEAFLTNNFDVTNEENDINHFNKVKAGICGDPQVEREFLCSTSDLLEERINELDSSE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PcyM_1438200 | 129 T | YILDTPKEV | 0.994 | unsp |