

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
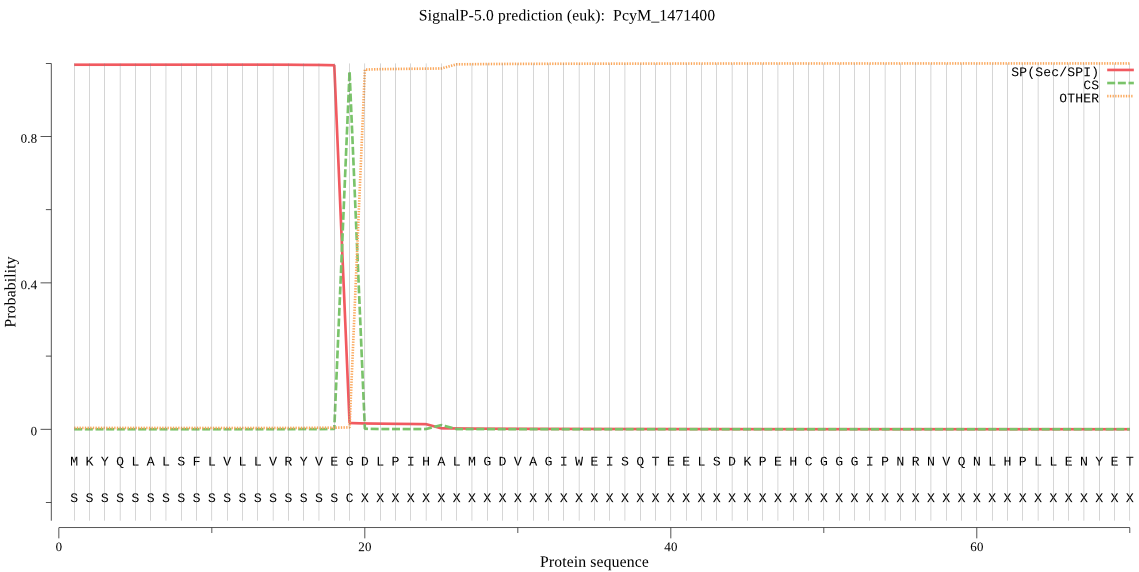
| PcyM_1471400 | SP | 0.002890 | 0.997002 | 0.000108 | CS pos: 19-20. VEG-DL. Pr: 0.9803 |
MKYQLALSFLVLLVRYVEGDLPIHALMGDVAGIWEISQTEELSDKPEHCGGGIPNRNVQN LHPLLENYETFLESNYGELETTSMKLTTEKINLSDKVNPRNNWTYLAVRDVKTNGIVGHW TMVYDEGFEVRVPGRRYFALFKYNRINSKRCPEPIETKDSTDSNCYMTDSTRTLIGWVLH ERVHKVHQNDKEKKVFQWGCFHGRKQEDVAVSSFVIHGAHGSRGGGETLAPKNQLSFAAI KQRSNYTKIRLSTGSPYGVPKTSLMSIYQRAREQIYGCRKQQSEEAKIRLTLPKAFSWGD PFSNNNFEEDVEDQMNCGSCYSIATLYSLHKRFEIWLLKKYKKKITMPRLSYQSILSCSP YNQGCDGGFPFLVGKQLYEFGIPTENSLPYGNSDSIKCEMSMGSYNNITSAKYKEEDLFF AAEYNYVSGCYECSNEYDMMNELFLNGPIVVAINATPQLLSLYKLGKQNGIYNTAMHENK VCDVPNEGFNGWQQTNHAVTIVGWGEEEKEEEQEGGNVLKYWVVRNTWGKEWGYKGYIKF QRGVNLAGIETQAVFLDPDLSRGRAGKISGQGGEGGEVGAESGSEGGDERGGKSGDERGG EGEAAMRPPIRHTPHAQRDNV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PcyM_1471400 | 297 S | PKAFSWGDP | 0.995 | unsp | PcyM_1471400 | 297 S | PKAFSWGDP | 0.995 | unsp | PcyM_1471400 | 297 S | PKAFSWGDP | 0.995 | unsp | PcyM_1471400 | 582 S | VGAESGSEG | 0.992 | unsp | PcyM_1471400 | 148 S | NRINSKRCP | 0.991 | unsp | PcyM_1471400 | 252 S | KIRLSTGSP | 0.996 | unsp |