

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
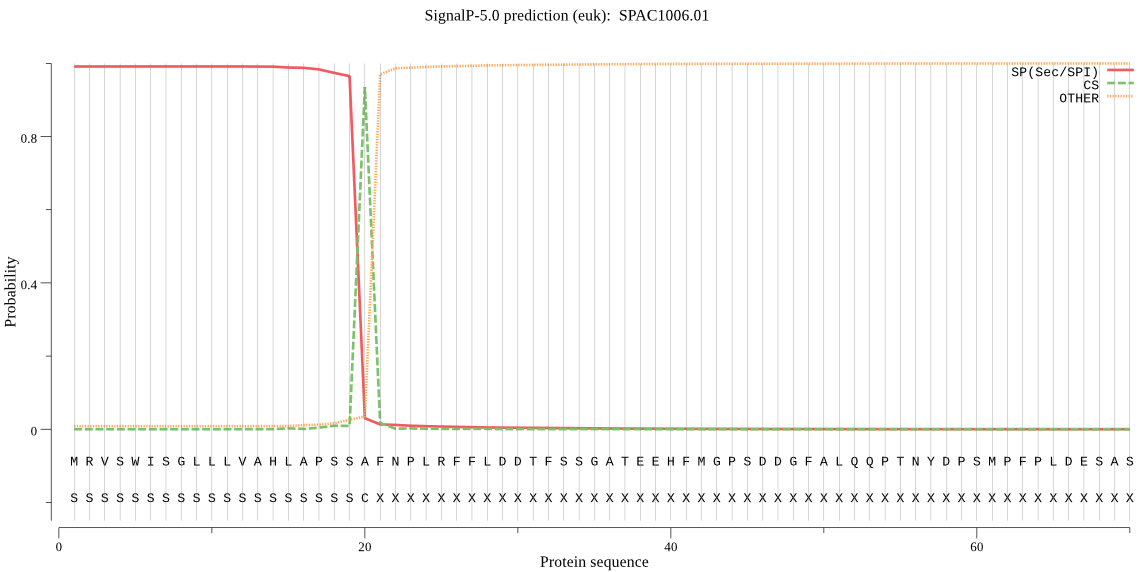
| SPAC1006.01 | SP | 0.004502 | 0.994645 | 0.000852 | CS pos: 20-21. SSA-FN. Pr: 0.9307 |
MRVSWISGLLLVAHLAPSSAFNPLRFFLDDTFSSGATEEHFMGPSDDGFALQQPTNYDPS MPFPLDESASAAVDAVSNNYIVMFKPSVDKSKLEQHHRWIEHLHEKRSLDFKDVSTFLMK HTFEIGDAFLGYAGRFSPWLVAELQKHPDIALVEPDRVMHVMTEQTFAPWGLARVSHRKK LGFFTMTRYQYNETAGEGVTAYVIDTGINIEHQDFQGRATWGATIPTGEGEVDDHGHGTH VAGTIAGKTFGVSKNAKLVAVKVMRADGTGTVSDIIKGIEFAFKQSKKDKESIASVVNMS IGGDASTALDLAVNAAIAGGLFFAVAAGNDAEDACGTSPARVSNAMTVGASTWNDQIASF SNIGSCVDIFAPGSLILSDWIGSNRASMILSGTSMASPHVAGLAAYFISLDPSLANHPVE LKKYMLKFALKDLLNGIPEDTPNVLAFNNYE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| SPAC1006.01 | 286 S | AFKQSKKDK | 0.996 | unsp |