

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC1296.03c | SP | 0.016171 | 0.982974 | 0.000855 | CS pos: 22-23. IHA-LP. Pr: 0.9717 |
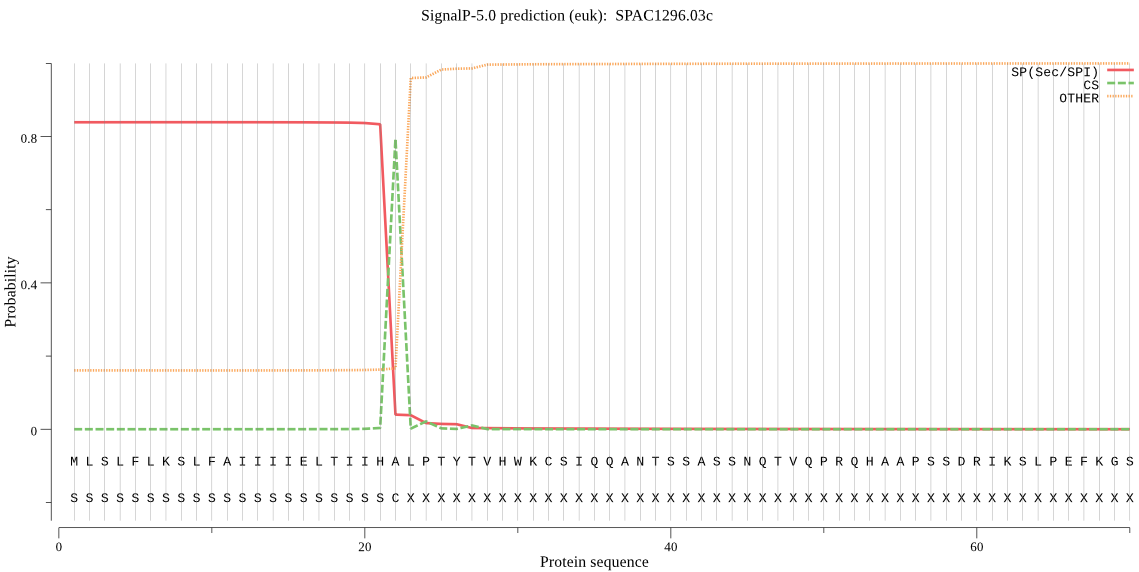
MLSLFLKSLFAIIIIELTIIHALPTYTVHWKCSIQQANTSSASSNQTVQPRQHAAPSSDR IKSLPEFKGSLPELYSGYLEANSDKSLFYTYAPAVVDSETFIVWLQGGPGCAGTLGFFSE NGPIEISQSSPSPSLNPESWTNFANMLWLDQPFGTGYSQGQAAYTTTIEEASSDFVNALK SFYQKFPHLMKKKLYLVGESYGSIWSANFAEALLSEPSLNINFMGVGIVSGLTADYETQE QITASIWVEHISKLGYYFNNTSSTISEEFKKRNKECQYDSVLNRLTFPTEQYPIWRPEYN FSTSTSLRKREALDGEDIGNVFNSISGCDLYSLSNFLLYLENSCVITYDVSLDCSFNEYN DPLITYLNREDVRSSLHATKASTALTSGEGVFADGCNFDLYKKIVSNNVESVLVEIIPRL TEKYKVSFLAGALDLQILWTGTLLALQNTTWNGWQGFTQSPGSLETTNGFTLDERNLAFT LSNSVGHMAPSKDPQMVREWLENTLLY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPAC1296.03c | 266 S | SSTISEEFK | 0.993 | unsp | SPAC1296.03c | 266 S | SSTISEEFK | 0.993 | unsp | SPAC1296.03c | 266 S | SSTISEEFK | 0.993 | unsp | SPAC1296.03c | 375 S | DVRSSLHAT | 0.995 | unsp | SPAC1296.03c | 63 S | DRIKSLPEF | 0.991 | unsp | SPAC1296.03c | 132 S | QSSPSPSLN | 0.993 | unsp |