

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC12G12.11c | OTHER | 0.999993 | 0.000001 | 0.000006 |
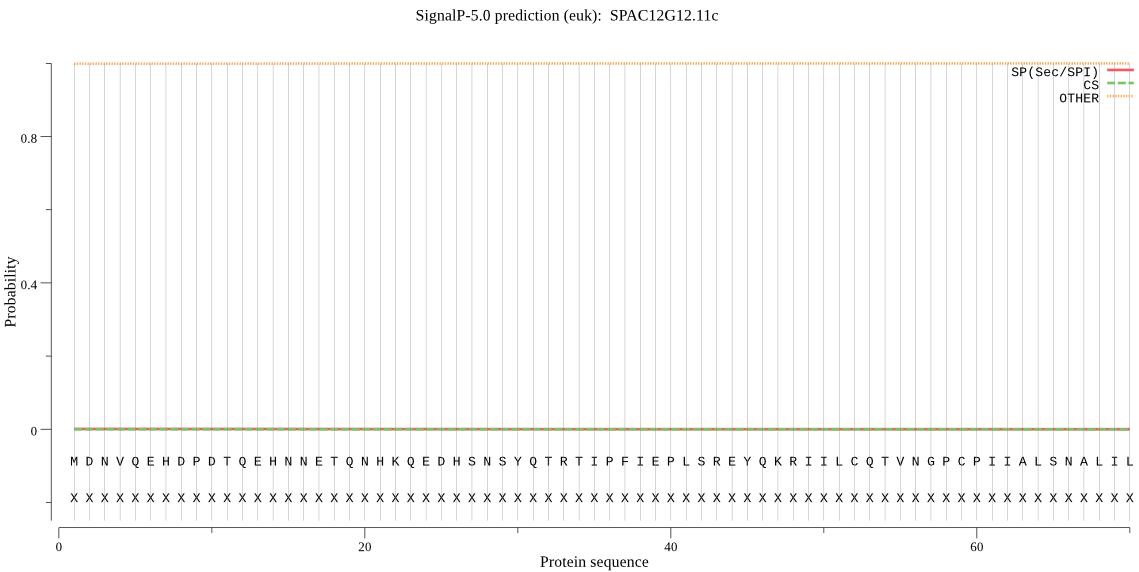
MDNVQEHDPDTQEHNNETQNHKQEDHSNSYQTRTIPFIEPLSREYQKRIILCQTVNGPCP IIALSNALILKSNVDRPFELPKKRYITPDELTEYLVEFAKAYGLCKNQQSLQDKLTSMHF GQQLNPCLYDIEKFEYGHEIFCTFGVRLVHGWILSDDMGLSDEDLSYLRKLEYYEKVADT FAERRSLLEMQEPLTEQQQDFLNNSTCVDKVMENRYTMQFLTNAGLKKILELVGPGEIVV VFRSSHFSTMYSNPDSFAQFTLVTDSGYARTGEDVVWETFDSQTVETGNGELCAANFIPA VYVLNQRKEEKKKRAKDDEQYAKRLAKEEEERGKKETPKKASNTPRRNKSNTQKSRKQSE NCLIS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPAC12G12.11c | 161 S | DMGLSDEDL | 0.995 | unsp | SPAC12G12.11c | 355 S | NTQKSRKQS | 0.997 | unsp |