

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| SPAC13A11.05 | mTP | 0.349343 | 0.053276 | 0.597381 | CS pos: 26-27. IPL-AT. Pr: 0.3746 |
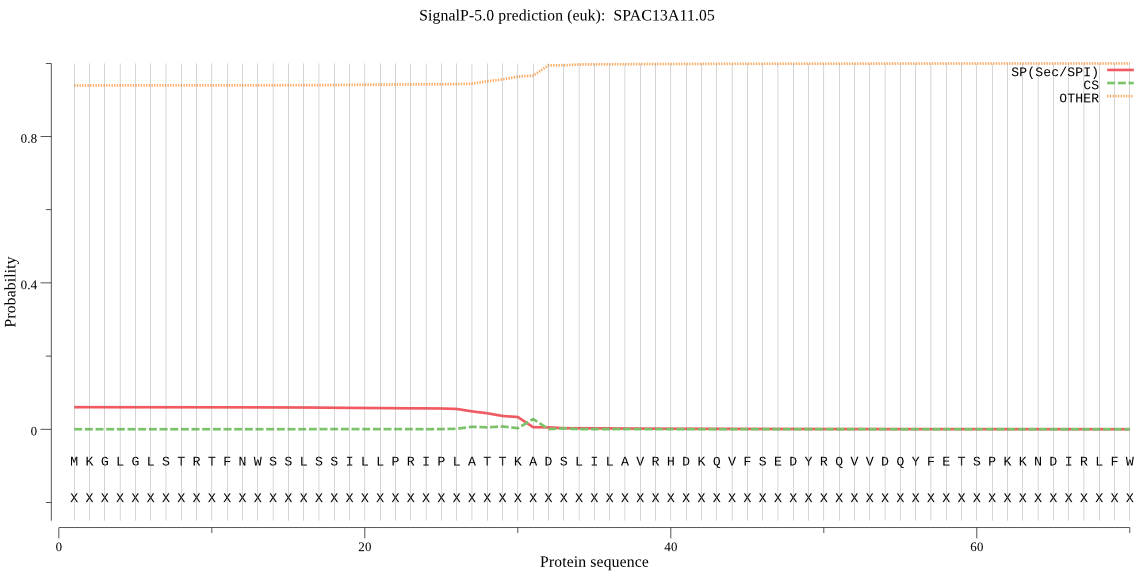
MKGLGLSTRTFNWSSLSSILLPRIPLATTKADSLILAVRHDKQVFSEDYRQVVDQYFETS PKKNDIRLFWNTQGFVRLAIVQLEENVSEKSVRSAAAEAAKILKSNGAKSIAVDGMGFPK DAALGAALATYDFSLRRDHLSVYQDEKVVEKENLFTSPAPERLTFQLLSNTSEKKTATAE ENAFKVGLIEAAAQNLARSLMECPANYMTSLQFCHFAQELFQNSSKVKVFVHDEKWIDEQ KMNGLLTVNAGSDIPPRFLEVQYIGKEKSKDDGWLGLVGKGVTFDSGGISIKPSQNMKEM RADMGGAAVMLSSIYALEQLSIPVNAVFVTPLTENLPSGSAAKPGDVIFMRNGLSVEIDN TDAEGRLILADAVHYVSSQYKTKAVIEASTLTGAMLVALGNVFTGAFVQGEELWKNLETA SHDAGDLFWRMPFHEAYLKQLTSSSNADLCNVSRAGGGCCTAAAFIKCFLAQKDLSFAHL DIAGVMDKQLNSWDCDGMSGRPVRTIIEVARKY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| SPAC13A11.05 | 60 S | YFETSPKKN | 0.994 | unsp | SPAC13A11.05 | 269 S | GKEKSKDDG | 0.992 | unsp |